File Menu: Difference between revisions
Jump to navigation
Jump to search
Created page with "Image:Spreadsheet_file_menu.png # '''New''' – Opens a blank spreadsheet.<br><br> # '''New from filtered''' - Transfers the rows visible under current filter conditions ..." |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image: | [[Image:Spreadsheet_file_menu_v2014.png]] | ||
<br> | |||
<ol> | |||
<li> | |||
{| | |||
|[[Image:File_menu_new.png]] | |||
| style="vertical-align:top; text-align:left;" | | |||
*'''New''' – Opens a blank spreadsheet. Note: if '''Force default template''' option in the '''Expert Panel''' is set by the administrator, the functionality of this command for a Non-Expert user will be identical to '''New from template'''<br><br> | |||
*'''New from Filtered''' - Transfers the rows visible under current filter conditions to a new spreadsheet project.<br><br> | |||
*'''New from Template''' - Opens a new spreadsheet containing all the columns defined in the currently selected default template (see '''Expert Panel''' for configuration options). The columns that were calculated by the program (properties or categories) will be automatically populated when adding new compounds. | |||
|} | |||
</li> | |||
<br> | |||
<li>'''Open''' - Opens any of the previously saved “Chemical Spreadsheet” (CHS) files.<br><br></li> | |||
<li> | |||
{| | |||
# '''Export''' - Allows export of the contents of the current spreadsheet to an SDF, RDF, TXT, CSV, or XLS formats. <br> '''NOTE:''' All the compounds in the active spreadsheet are exported, regardless of any filters applied at the moment of data export making some of the rows invisible.<br /> | |[[Image:File_menu_save.png]] | ||
| style="vertical-align:top; text-align:left;" | | |||
*'''Save''' - Saves the active spreadsheet with its current filename and location on the hard drive.<br><br> | |||
*'''Save As''' - Saves a copy of the active spreadsheet under the user-defined filename and location on the hard drive.<br><br> | |||
*'''Save Template''' - Saves the list of columns (names and definitions) in the active spreadsheet as a spreadsheet template file under the user-defined filename and .chstpl extension. | |||
|} | |||
</li> | |||
<br> | |||
<li>'''Close''' - Closes the active spreadsheet. A message box will prompt the user to save the active project.</li><br> | |||
<li> | |||
{| | |||
|[[Image:File_menu_import.png]] | |||
| style="vertical-align:top; text-align:left;" | | |||
*'''Import''' - Imports data to the active spreadsheet. The supported file formats include single structure files (CDX, MOL, SK2, and SKC), multiple structures files (SDF, RDF), as well as tab-delimited text (TXT) or comma separated value (CSV) files with chemical structures as SMILES strings. <br> '''NOTE:''' Importing new data to the open spreadsheet will append all the structures from a new data source. All the columns in the original spreadsheet associated with modules available in '''ACD/Percepta''' (i.e., the ones calculated from within the program) will be automatically populated with the calculated corresponding property values for the new appended molecules. <br><!--''See also [[Starting#How to import data into a spreadsheet?|How to import data into a spreadsheet?]]''--><br> | |||
*'''Import from Project''' - Imports data to the active spreadsheet from another project. After selecting this menu item a [[#Import_from_Project|dialog box]] is displayed guiding you through all steps of the import process. | |||
|} | |||
</li> | |||
<br> | |||
<li>'''Export''' - Allows export of the contents of the current spreadsheet to an SDF, RDF, TXT, CSV, or XLS formats. <br> '''NOTE:''' All the compounds in the active spreadsheet are exported, regardless of any filters applied at the moment of data export making some of the rows invisible.<br /></li> | |||
</ol> | |||
<br /> | <br /> | ||
==Import from Project== | |||
<ol> | |||
<li>Select the project to be used as a source of data for import from a dropdown box.<br> | |||
'''NOTE:''' Only those projects that are currently opened in '''ACD/Percepta''' are displayed in this list. | |||
<br> | |||
[[Image:import_project_step_1.png]] | |||
</li> | |||
<br> | |||
<li>Drag all columns of the source project that you wish to be imported to the right-hand side pane. | |||
<br> | |||
[[Image:import_project_step_2.png]] | |||
</li> | |||
<br> | |||
<li>Link matching columns between the source and destination projects. If both projects contain columns with identical names, these will be linked automatically. Otherwise, if certain columns contain the same kind of data, but are named differently, you will need to link them manually. | |||
<br> | |||
[[Image:import_project_step_3.png]] | |||
</li> | |||
<br> | |||
<li>Specify whether the program should simply append the entries from the source project without any checking for possible duplicates, or merge the data from both source and destination projects with matching entries identified either by comparing values stored in a particular column or by molecular structure. In the latter case, a honeycomb diagram is displayed illustrating whether the central molecule would match (denoted by "=" sign) the molecules on the outside under the currently selected settings. | |||
<br> | |||
[[Image:import_project_step_4.png]] | |||
</li> | |||
<br> | |||
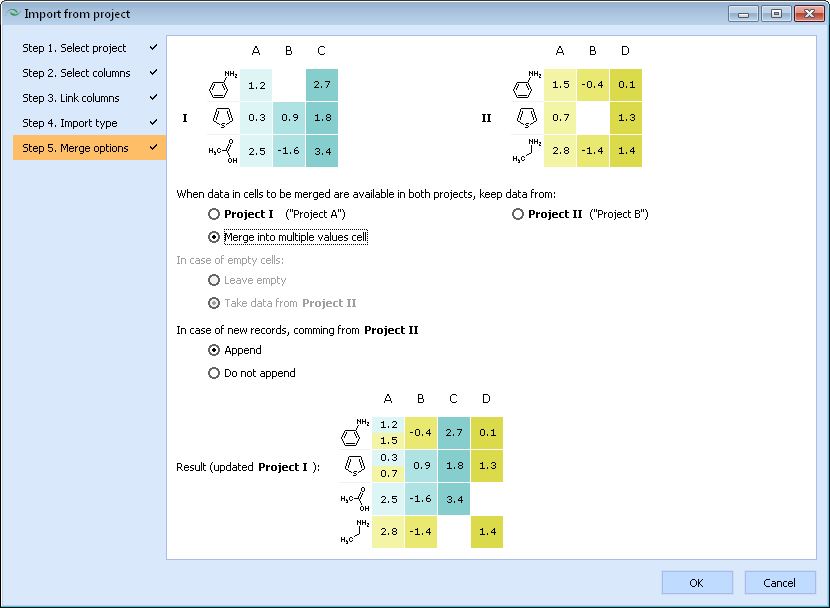
<li>Finally, if "Merge" import type was selected on step 4, several additional options specifying how to deal with missing or overlapping values need to be set. This window also presents a schematic diagram illustrating how the resulting project would look like if the example data shown above would be merged under the currently selected settings. | |||
<br> | |||
[[Image:import_project_step_5.png]] | |||
</li> | |||
</ol> | |||
Revision as of 14:50, 12 December 2014
- Open - Opens any of the previously saved “Chemical Spreadsheet” (CHS) files.
- Close - Closes the active spreadsheet. A message box will prompt the user to save the active project.
-
- Import - Imports data to the active spreadsheet. The supported file formats include single structure files (CDX, MOL, SK2, and SKC), multiple structures files (SDF, RDF), as well as tab-delimited text (TXT) or comma separated value (CSV) files with chemical structures as SMILES strings.
NOTE: Importing new data to the open spreadsheet will append all the structures from a new data source. All the columns in the original spreadsheet associated with modules available in ACD/Percepta (i.e., the ones calculated from within the program) will be automatically populated with the calculated corresponding property values for the new appended molecules. - Import from Project - Imports data to the active spreadsheet from another project. After selecting this menu item a dialog box is displayed guiding you through all steps of the import process.
- Import - Imports data to the active spreadsheet. The supported file formats include single structure files (CDX, MOL, SK2, and SKC), multiple structures files (SDF, RDF), as well as tab-delimited text (TXT) or comma separated value (CSV) files with chemical structures as SMILES strings.
- Export - Allows export of the contents of the current spreadsheet to an SDF, RDF, TXT, CSV, or XLS formats.
NOTE: All the compounds in the active spreadsheet are exported, regardless of any filters applied at the moment of data export making some of the rows invisible.
Import from Project
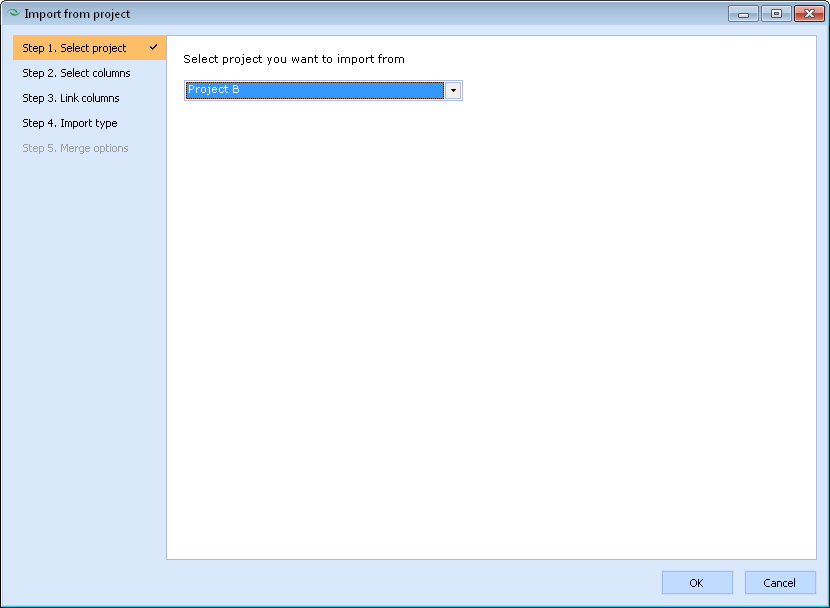
- Select the project to be used as a source of data for import from a dropdown box.
NOTE: Only those projects that are currently opened in ACD/Percepta are displayed in this list.
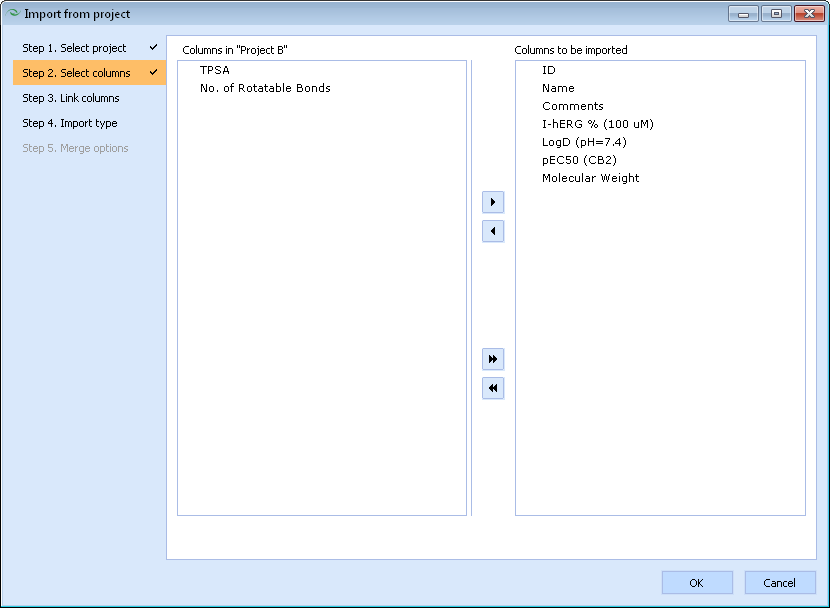
- Drag all columns of the source project that you wish to be imported to the right-hand side pane.
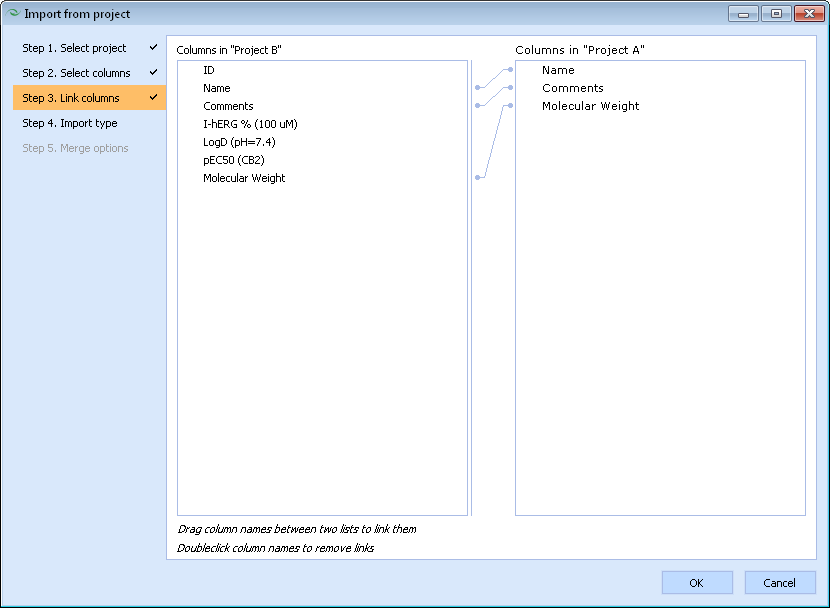
- Link matching columns between the source and destination projects. If both projects contain columns with identical names, these will be linked automatically. Otherwise, if certain columns contain the same kind of data, but are named differently, you will need to link them manually.
- Specify whether the program should simply append the entries from the source project without any checking for possible duplicates, or merge the data from both source and destination projects with matching entries identified either by comparing values stored in a particular column or by molecular structure. In the latter case, a honeycomb diagram is displayed illustrating whether the central molecule would match (denoted by "=" sign) the molecules on the outside under the currently selected settings.
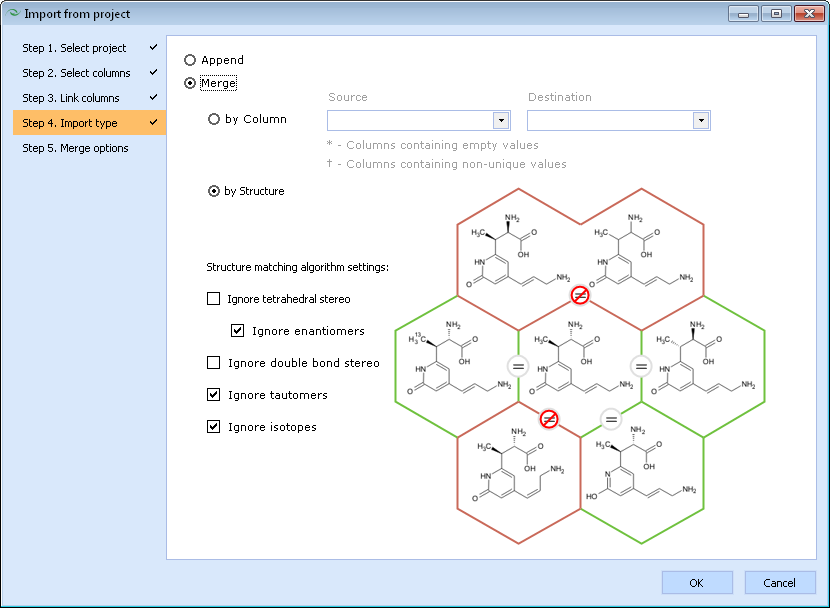
- Finally, if "Merge" import type was selected on step 4, several additional options specifying how to deal with missing or overlapping values need to be set. This window also presents a schematic diagram illustrating how the resulting project would look like if the example data shown above would be merged under the currently selected settings.