Special compound classes
Inorganic compounds
Properties will NOT be calculated for inorganic compounds. Inorganic compound – a compound with no carbon atoms in it.
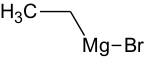
Metalo-organic compounds
Properties will NOT be calculated for metallo-organic compounds. Metallo-organic compound – a compound with a covalent bond between metal and non-metal atoms. All elements are considered metals except: H, B, C, N, O, F, Si, P, S, Cl, Ge, Ar, Se, Br, Te, I, Sn, Sb, Bi.
The following are examples of compounds that will be treated as metallo-organic (no properties will be calculated for them):
NOTE:
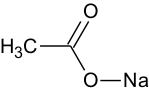
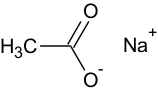
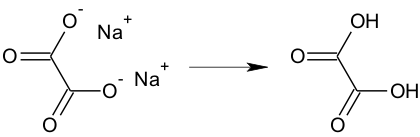
Salts, e.g. sodium acetate, will be calculated if it is drawn in the ionic form as shown below.
Salts, hydrates and mixtures
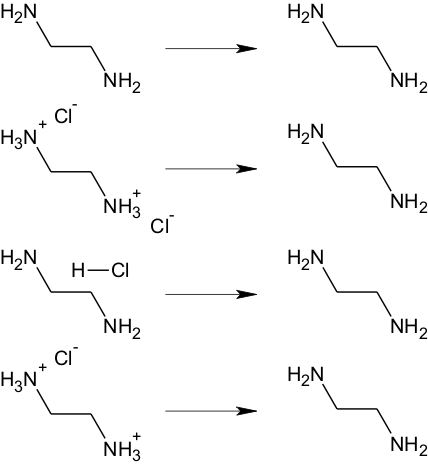
Salts
For all of the modules (except Skin/Eye irritation) salts are converted into the neutral form (where possible) before any calculation. The rationale is that in the absolute majority of the properties calculated by the corresponding software products happens in buffer conditions, i.e. it doesn’t matter whether compound is administered as free acid/base or salt – it will have the same proportion of ionized and unionized forms, which depends solely on ionization properties of free acid/base and pH of the environment. The only exception is Skin/Eye irritation, where buffering is non-existent.
The following compounds will produce the same predictions as they are converted to the “free” ethyl diamine first.
Hydrates
Water molecules are removed from non-covalent hydrates before any prediction.
Mixtures
For mixtures, only the biggest organic component of the mixture will be used for the prediction.
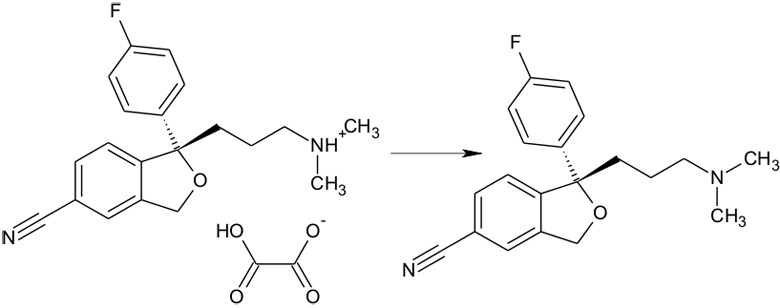
This includes salts with organic counter ions, as shown below.
NOTE:
For a mixture, each compound is checked separately to see if it is inorganic or metallo-organic.
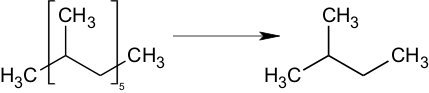
Stereoisomers
Stereoisomer centers of any kind (R/S, D/L, cis/trans, etc.) are NOT considered in the predictions. All stereoisomers will produce the same prediction.
Proteins
Properties will be calculated for proteins and polypeptides. However we recommend that the MW not surpass 2000 Daltons. Since the internal training libraries do contain only some small proteins and polypeptides, any conformational effects will NOT be taken into account.
Polymers
Properties will be calculated for polymers when they are represented in a conventional form as any other organic compound. However, since the internal training libraries do not contain any polymers, we recommend treating such predictions with caution as they are highly likely to be unreliable due to excessive extrapolation.
Polymer structures drawn using brackets to indicate repeating motifs will NOT be recognized as polymers and the brackets will be omitted when pasting such molecules into the program.
High Molecular Weight Compounds
Properties will be calculated for high MW compounds. We recommend that the MW not surpass 2000 Daltons (even more conservative threshold would be 1500 Daltons) as conformational and supramolecular effects will NOT be taken into account. Calculation time will increase with larger compounds.