Classic Training Workspace: Difference between revisions
Jump to navigation
Jump to search
The first version of the page |
mNo edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 14: | Line 14: | ||
# The main '''Training panel''' consists of several expandable-collapsible sections for different physicochemical properties | # The main '''Training panel''' consists of several expandable-collapsible sections for different physicochemical properties | ||
# '''ACD LogP/LogD''' and '''ACD Solubility''' sections may display training data for ACD/LogP Classic and ACD/Solubility Classic algorithms from legacy PCD files in read-only mode. In recent versions of Percepta, training LogP and Solubility calculations is recommended via the respective [[Training|GALAS algorithms]] | # '''ACD LogP/LogD''' and '''ACD Solubility''' sections may display training data for ACD/LogP Classic and ACD/Solubility Classic algorithms from legacy PCD files in read-only mode. In recent versions of Percepta, training LogP and Solubility calculations is recommended via the respective [[Training|GALAS algorithms]] | ||
<br> | |||
[[Image: | [[Image:Classic_training_pKa_database.png]] | ||
# Switch to '''pKa Database''' section to perform pKa training | # Switch to '''pKa Database''' section to perform pKa training | ||
| Line 22: | Line 22: | ||
# Click on a table row to edit the respective entry | # Click on a table row to edit the respective entry | ||
[[Image:Classic_training_edit_record.png| | ===Edit record dialog=== | ||
[[Image:Classic_training_edit_record.png|right]] | |||
# Enter the experimental pKa value and optionally, fill other related data fields, such as confidence limits, temperature, ionic strength, literature reference and any custom notes | # Enter the experimental pKa value and optionally, fill other related data fields, such as confidence limits, temperature, ionic strength, literature reference and any custom notes | ||
Latest revision as of 13:53, 20 June 2025
Overview
Classic Training is a specialized workspace that is used to train ACD/pKa Classic prediction model with user-defined experimental data.
Classic training works with PhysChem Database (PCD) format files, therefore, prior to using this workspace, a suitable database file should be created or loaded via the Data Source panel and if necessary, populated with chemical structures to be used in training via Spreadsheet View.
Interface
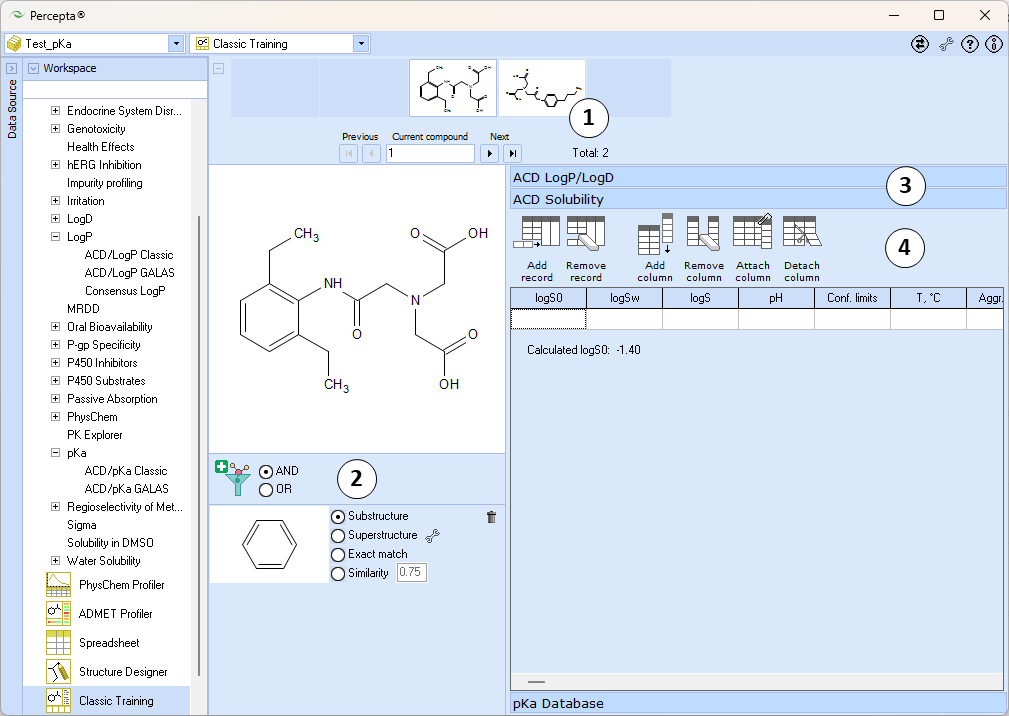
- Standard Navigation bar is used to browse the structures included in the database. Unlike Prediction modules, here it does not support addition of new structures to the project
- Filter panel allows applying structural filters analogous to Spreadsheet View directly from the workspace
- The main Training panel consists of several expandable-collapsible sections for different physicochemical properties
- ACD LogP/LogD and ACD Solubility sections may display training data for ACD/LogP Classic and ACD/Solubility Classic algorithms from legacy PCD files in read-only mode. In recent versions of Percepta, training LogP and Solubility calculations is recommended via the respective GALAS algorithms
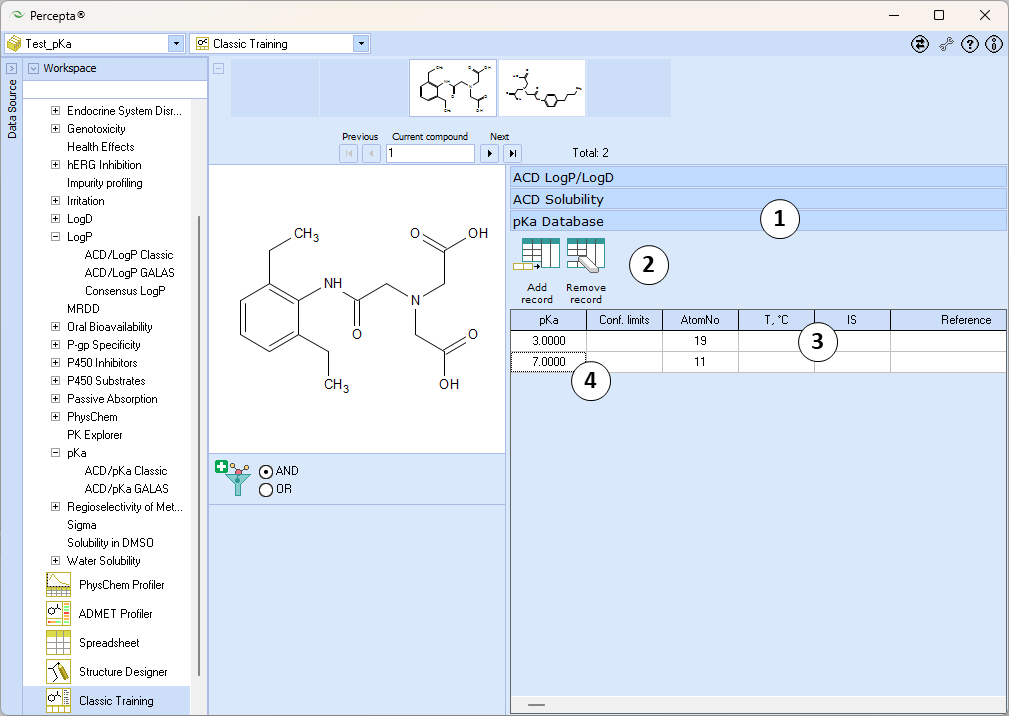
- Switch to pKa Database section to perform pKa training
- Use the toolbar buttons to add a new pKa entry or to remove the selected entry from the data table
- The data table displays pKa data currently entered in the database for the active compound
- Click on a table row to edit the respective entry
Edit record dialog
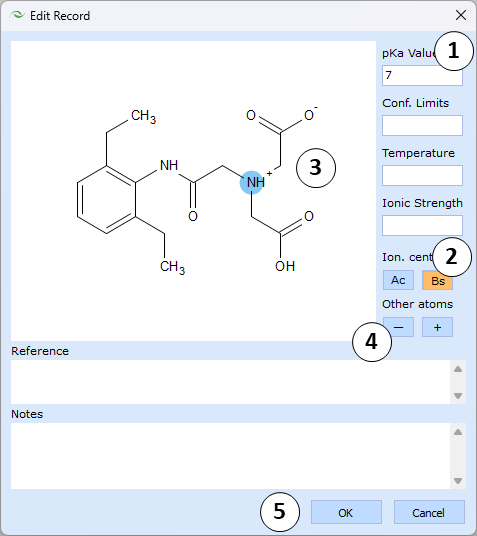
- Enter the experimental pKa value and optionally, fill other related data fields, such as confidence limits, temperature, ionic strength, literature reference and any custom notes
- Select the atom that loses the proton during the characterized dissociation stage. For this purpose, mark "Ac" or "Bs" buttons to specify acidic or basic ionization, respectively, and then click on the corresponding atom in the Structure panel
- The selected atom gains a color highlight: red for acidic center, blue for basic one
- In order to specify the exact ionic form of the compound at relevant pH, mark "-" or "+" buttons and click on atoms in the Structure panel to assign negative or positive charges
- Finally, click OK to confirm the edits or Cancel to discard them