ADMET Profiler
Drug Profiler is an integrated workspace that aggregates the output of various PhysChem, ADME, and Drug safety-related predictors included in ACD/Percepta and presents the data as the compound's overall ADME/Tox profile with individual predictions categorized according to user-adjustable thresholds and visualized using color-coded indications. Drug Profiler enables the researcher to identify the most suitable structures that meet specific criteria for a wide range of properties:
- PhysChem
- LogP
- Solubility
- H-bonding
- Molecular size and flexibility
- Drug- and Lead-likeness rules
- ADME
- Human Intestinal Absorption (HIA) and Caco-2 permeability
- Plasma protein binding (PPB)
- Blood-brain pernetration and accessibility to CNS
- Metabolic stability
- Drug Safety
- P-gp substrate specificity
- Cytochrome P450 inhibitor specificity
- hERG inhibitor specificity
- Mutagenicity in Ames test
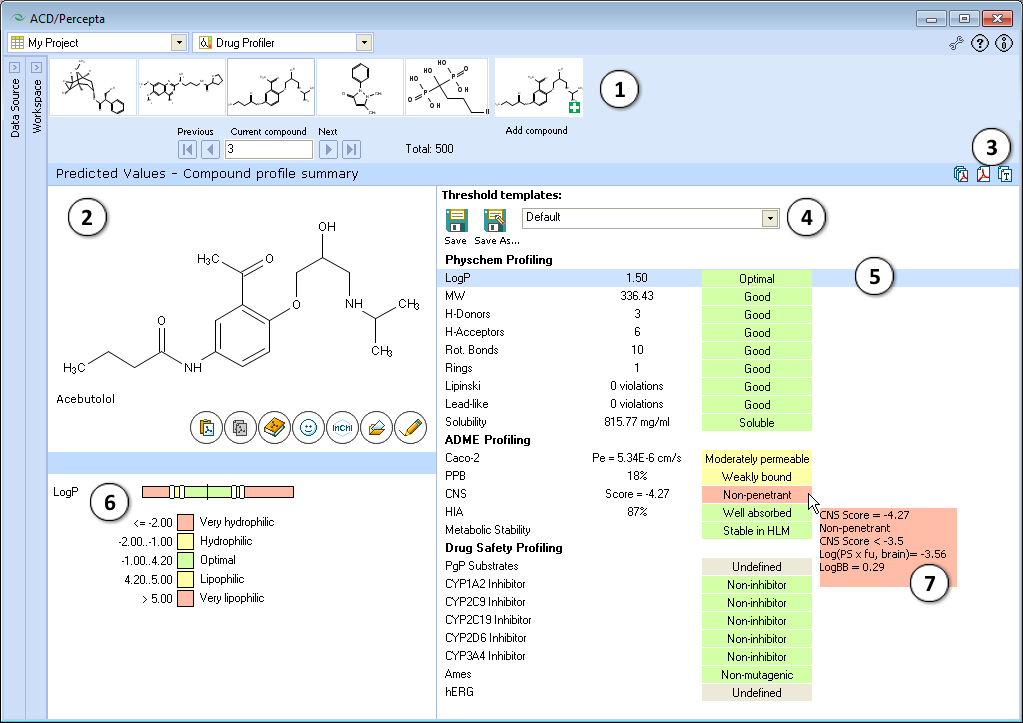
- Navigation Bar
The Navigation Bar appears in all workspaces except the Spreadsheet View. It displays presented structure and two previous as well as two next stuctures in the corresponding dataset. If no dataset is selected, the Navigation Bar displays the history of performed calculations.
When the compound outside of the dataset is used as an input structure for calculation, the user can add it to the current dataset:
- Click the
 pictogram to replace the current structure with the new one. All subsequent calculations will be performed automatically for the added compound.
pictogram to replace the current structure with the new one. All subsequent calculations will be performed automatically for the added compound. - Click the
 pictogram to add new compound to the current dataset. It will appear as the last record in the dataset and all subsequent calculations will be performed automatically.
pictogram to add new compound to the current dataset. It will appear as the last record in the dataset and all subsequent calculations will be performed automatically. - The structure browse bar displays the active structure number and the total number of records in the current dataset. Scroll through the database using the browse buttons: First/Last record, Previous/Next record or pressing the up/down arrow keys on the keyboard to go to the next/previous record. Type a record number to view it directly.
- Click the
- Structure Pane
The Structure Pane displays a 2-D representation of the chemical structure used for calculation. A compound name is displayed only for those structures which are found in the Dictionary. (Ctrl+V): Pastes an input structure from the clipboard to the Structure Pane. Also, if you are pasting a SMILES string from the clipboard, you can save a step by clicking directly on this button to display the 2-D structure. The calculation will be performed automatically for the input structure.
(Ctrl+V): Pastes an input structure from the clipboard to the Structure Pane. Also, if you are pasting a SMILES string from the clipboard, you can save a step by clicking directly on this button to display the 2-D structure. The calculation will be performed automatically for the input structure. (Ctrl+C): Copies the active structure in the Structure Pane to the clipboard.
(Ctrl+C): Copies the active structure in the Structure Pane to the clipboard. (Ctrl+D): Opens the Dictionary with >117,000 available names to find the corresponding structure and display it. When a compound is selected from the list and the selection confirmed by clicking OK, calculations for the chosen compound will be performed automatically.
(Ctrl+D): Opens the Dictionary with >117,000 available names to find the corresponding structure and display it. When a compound is selected from the list and the selection confirmed by clicking OK, calculations for the chosen compound will be performed automatically. (Ctrl+S): Type or paste SMILES notation string in this field. The string will automatically be converted to 2-D structure. Calculation will be performed automatically for the input structure.
(Ctrl+S): Type or paste SMILES notation string in this field. The string will automatically be converted to 2-D structure. Calculation will be performed automatically for the input structure.
 (Ctrl+O): Opens a file containing a molecular structure. ACD/Percepta supports all versions of MDL *.mol files, ACD/ChemSketch *.sk2, as well as ISIS Draw *.skc, and ChemDraw *.cdx formats.
(Ctrl+O): Opens a file containing a molecular structure. ACD/Percepta supports all versions of MDL *.mol files, ACD/ChemSketch *.sk2, as well as ISIS Draw *.skc, and ChemDraw *.cdx formats. (Ctrl+E): Allows to edit the active molecule.
(Ctrl+E): Allows to edit the active molecule.  Creates a PDF report with the displayed results.
Creates a PDF report with the displayed results. Copies the displayed results to the clipboard.
Copies the displayed results to the clipboard.- The dropdown list of available threshold templates. Use default thresholds or select any of the previously saved templates from the dropdown, and the property categories will be automatically re-assigned to match the newly selected set of thresholds. If any of the thresholds are altered compared to last saved version, this is indicated by an asterisk displayed next to the name of the template. The changes can be saved under the same template name, by pressing Save button, or alternatively, transfered to a new template by choosing Save as... command.
- A list of properties included in the profile, their calculated values, and category indication with a short textual description. The property list is interactive - click the property of interest to display its value scale and view or edit the cut-offs separating the categories in the lower left part of the window.
- Displays the value scale of corresponding property and the list of cut-offs defining different categories. A black vertical line indicates the position of the current compound on the property scale. Move the sliders to alter the numerical cut-offs separating the categories.
- Hover over the property value to view a screen-tip with more details regarding the particular prediction. For some properties, the screen-tips display additional quantitative parameters of relevance (e.g., CNS), predicted probabilities of positive assay outcome in case of probabilistic properties, and/or the numerical conditions that the compound had fulfilled in obtain to be classified in a particular category.