ACD/LogS0 GALAS
Overview
This module predicts intrinsic solubility (LogS0, mmol/ml) of a compound in water at 25°C using a set of >6,800 compounds.
Features
- Calculates a Reliability Index for every prediction
- Performs a similarity search and displays top 5 most similar structures from the training sets of the model
Interface
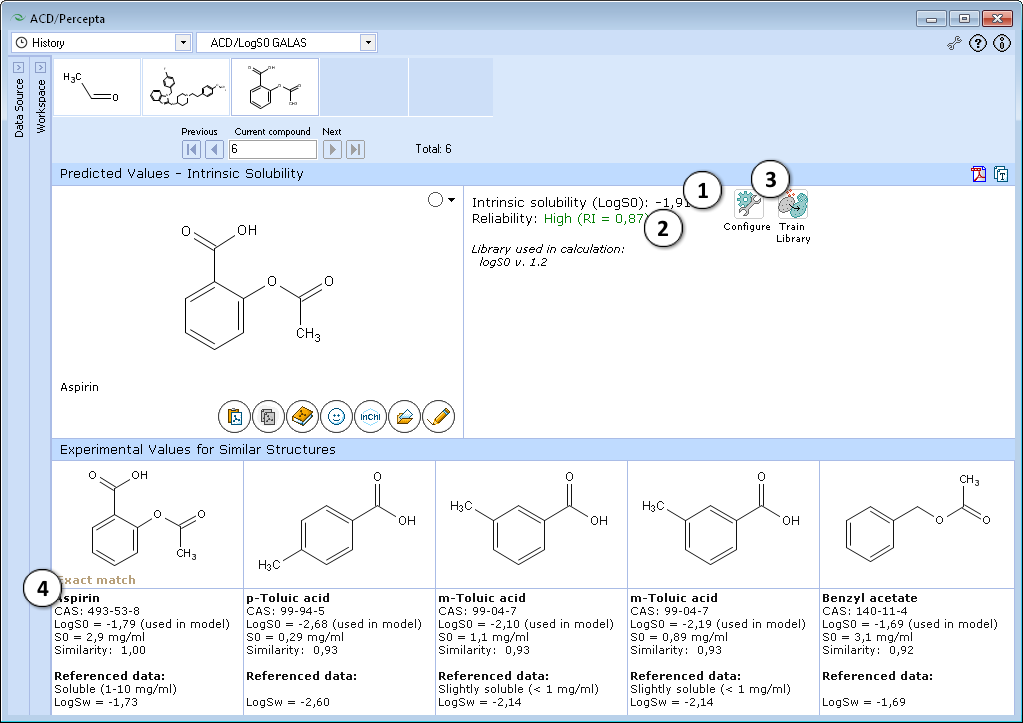
- Quantitative estimate of the compound's intrinsic solubility in water
- Indication of the prediction reliability along with the Reliability Index value
- "Configure" and "Train" buttons provide the means to select the training library for use in calculations and to add new data to that library. The name of the currently selected library is indicated with italic font.
- Up to 5 similar structures from the training set with experimental values
Note: Prediction reliability classification according to Reliability Index (RI) values:
- RI < 0.3 – Not Reliable,
- RI in range 0.3-0.5 – Borderline Reliability,
- RI in range 0.5-0.75 – Moderate Reliability,
- RI >= 0.75 – High Reliability
Technical information
Description of ACD/LogS0 GALAS Algorithm
ACD/LogS0 GALAS module provides provides the quantitative estimate of the compound’s solubility in water at 25°C (in terms of intrinsic solubility Log S0, mmol/ml) derived on the basis of GALAS (Global, Adjusted Locally According to Similarity) modeling methodology (please refer to [1] for more details).
Each GALAS model consists of two parts:
- Global (baseline) statistical model that reflects general trends in the variation of the property of interest.
- Similarity-based routine that performs local correction of baseline predictions taking into account the differences between baseline and experimental logS0 values for the most similar training set compounds.
GALAS methodology also provides the basis for estimating reliability of predictions by the means of calculated Reliability Index (RI) value that takes into account:
- Similarity of tested compound to the training set molecules.
- Consistence of experimental logS0 values and baseline model prediction for the most similar similar compounds from the training set.
Reliability Index ranges from 0 to 1 (0 corresponds to a completely unreliable, and 1 - a highly reliable prediction) and serves as an indication whether a submitted compound falls within the Model Applicability Domain. Compounds obtaining predictions RI < 0.3 are considered outside of the Applicability Domain of the model.
Experimental Data
Training set size: 4,764
Internal validation set size: 2,043
Main sources of experimental data:
- Reference books:
- The Merck Index. An Encyclopedia of Chemicals, Drugs, and Biologicals, O'Neil, M.J., Smith, A., Heckelman, P.E., Budavari, S., Eds. 13th Edition, Merck & Co., Inc., Whitehouse Station, NJ, 2001
- Therapeutic Drugs, Dolery, C., Ed. 2nd Edition, Churchill Livingstone, New York, NY, 1999
- Clarke's Isolation and Identification of Drugs, Moffat, A.C., Jackson, J.V., Moss, M.S., Widdop, B., Eds. 2nd Edition, The Pharmaceutical Press, London, 1986
- Various articles from peer-reviewed scientific journals*
* - Articles reporting the models of solubility in pure water (LogSw) by other authors were the predominant type among analyzed literature, meaning that each publication contained larger collections of experimental data (usually in the order of tens or hundreds compounds) compiled from corresponding original experimental articles. Original LogSw data had been converted to LogS0 prior to modeling.
Internal Validation
| Subset | Coverage of the entire internal validation set (N=2,043) |
R2 | RMSE | ||
|---|---|---|---|---|---|
| RI > 0.3 N = 1,990 |
|
0.83 | 0.84 | ||
| RI > 0.5 N = 1,663 |
|
0.87 | 0.77 | ||
| RI > 0.75 N = 567 |
|
0.93 | 0.64 |