P-gp Inhibitors
Overview
P-glycoprotein (P-gp) is a clinically relevant efflux transporter that extrudes compounds from a large variety of cells. Its function has been associated with the drugs’ absorption, distribution, excretion, CNS effects, multidrug resistance (MDR). P-gp transports a variety of natural compounds and drugs of different therapeutic areas.
Rapid identification of drug candidates that are P-gp substrates and/or inhibitors is possible using P-gp specificity module. Filtering and exclusion of P-gp substrates/inhibitors from huge ‘in-house’ libraries of synthesized compounds or virtual libraries is possible, followed by exclusion of such compounds from further development. P-gp specificity module may serve as an initial screen that could replace screening test based on P-gp ATPase activity measurements and partially replace expensive experiments with P-gp expressing cell monolayers and P-gp knock-out animals.
Training of P-gp specificity models with ‘in-house’ data allows producing reliable predictions of P-gp interaction with compounds synthesized in your company.
Features
- Calculates probabilities of the analyzed compound being a P-gp substrate or inhibitor.
- Calculates Reliability Index (RI values) of predictions that show whether tested compounds belong to Applicability Domain of predictive model?
- In some cases the explanation why tested compound can be P-gp substrate or inhibitor is added (supplementary classification P-gp specificity models).
- Displays experimental values for similar compounds (a fully browsable P-gp specificity database is supplemented).
- Training of models using ‘in-house’ data is possible.
- Training set of >1,000 compounds for P-gp substrate algorithm, and of >1,500 compounds for P-gp inhibitor algorithm was used
- Two models of P-gp substrate and inhibitor specificity were build - classification and probabilistic
- The classification models, used P-gp substrate and inhibitor specificity rules, based on ionization, molecular size and biological class of compounds (analogs of peptides, alkaloids, anthracyclines, etc.)
- Statistical algorithms calculate the probabilities of compound being P-gp substrate or inhibitor and estimate the reliability of every prediction by means of Reliability Index calculation (see more information about Trainable Models)
- Classification based on the experimental data is presented for similar structures
- Reference data were compiled from more than 800 original publications
Interface
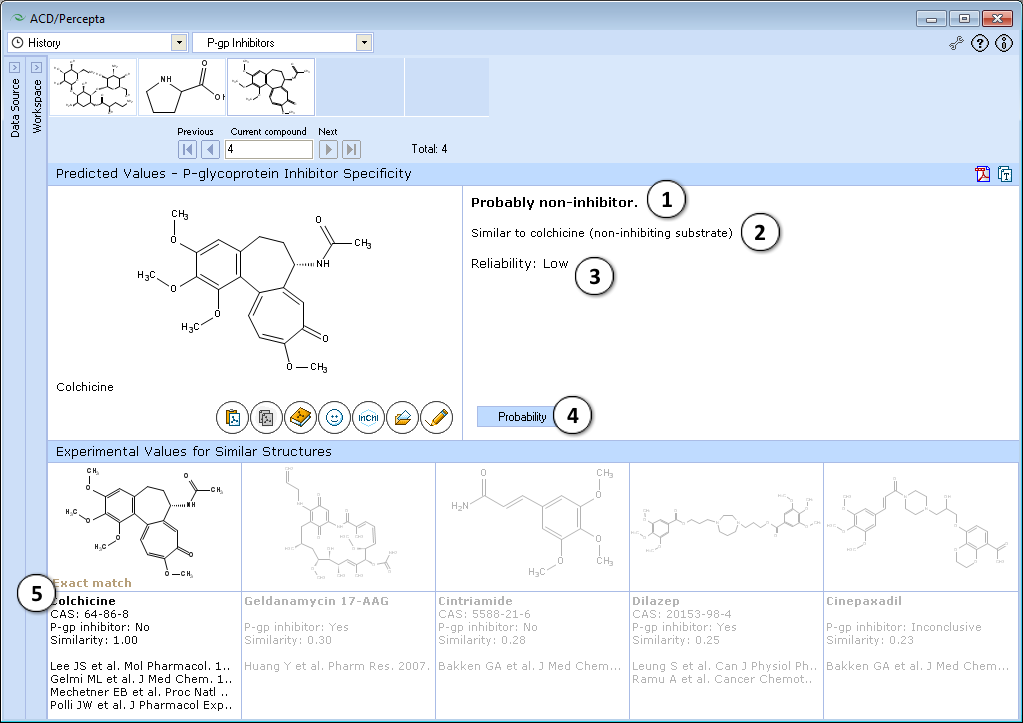
- Classification of compounds as inhibitors or non-inhibitors
- Description of prediction
- Reliability of prediction (low, medium, high)
- Switch to the probabilistic model
- Up to 5 most similar structures in the P-gp DB and references
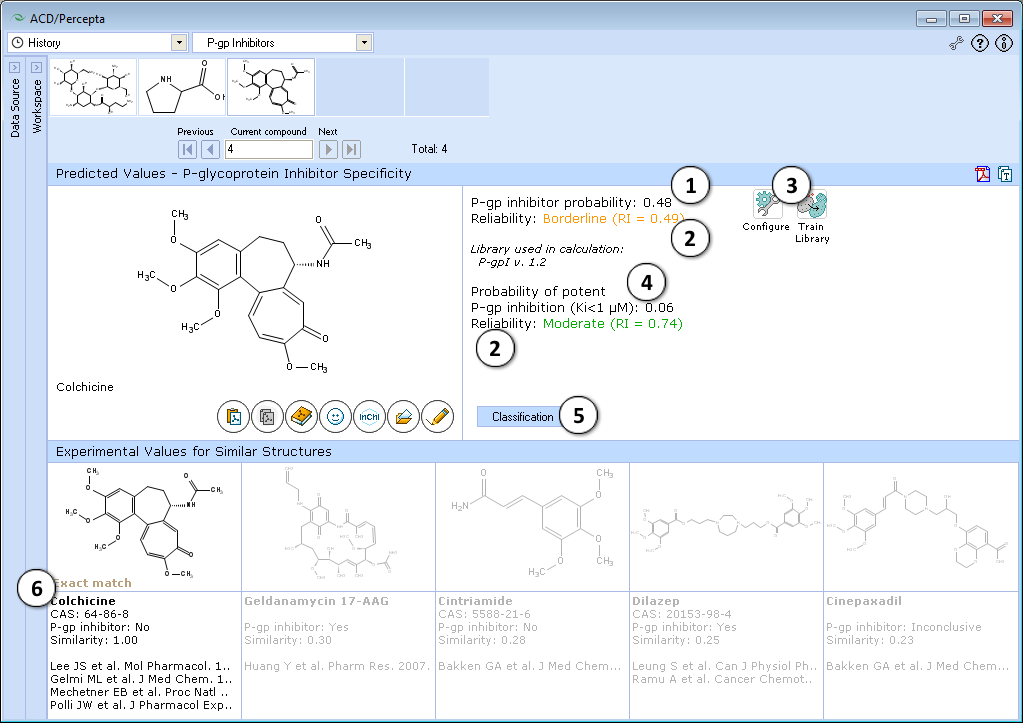
- Probability, ranging from 0 (definite non-inhibitor) to 1 (definite inhibitor)
- Indication of the P-gp Inhibitor probability prediction reliability along with the Reliability Index value
- Probability between 0 (definitely not a high-affinity inhibitor) and 1 (definitely a high-affinity inhibitor)
- Indication of the potent P-gp inhibition probability prediction reliability along with the Reliability Index value
- Switch to the knowledge based C-SAR model
- Up to 5 most similar structures in the P-gp DB and references
Note: Prediction reliability classification according to Reliability Index (RI) values:
- RI < 0.3 – Not Reliable,
- RI in range 0.3-0.5 – Borderline Reliability,
- RI in range 0.5-0.75 – Moderate Reliability,
- RI >= 0.75 – High Reliability
- Switch to P-gp Specificity\P-gp Inhibitor module.
- P-gp inhibitor specificity module is organized in the same way as Substrate specificity module. Classification of compounds as inhibitors or non-inhibitors along with text comments and reliability estimates is displayed by default. As could be seen from the screenshot, colchicine is classified as P-gp non-inhibitor.
- Switch between knowledge-based and probabilistic predictors.
- Probabilistic predictor provides two statistical models that estimate whether the test compound inhibits P-gp-mediated efflux, and whether it is a potent inhibitor with Ki < 1 μM. Both predicted probabilities are supplemented by RI values.
- Up to five most similar structures from P-gp DB are shown with experimental values and references.
Technical information
Experimental data
There are many in vitro and in vivo tests used in P-gp specificity studies that often produce contradictive results. P-gp specificity model is based on the data collected from scientific literature. The following assays for substrates were considered:
- In vitro polarized transport across P-gp expressing cell monolayers
- In vivo BBB models with P-gp knock-out animals, P-gp mediated drug resistance.
The respective assays for inhibitors were as follows:
- Drug efflux inhibition across/out of P-gp expressing cells
- MDR reversion.
Overall data set for P-gp substrates contains >1000 compounds, for inhibitors >1500 compounds.
Reference database
P-gp Specificity\P-gp DB module contains a browsable database of 2,290 compounds with experimental data related to their interactions with P-gp. Each compound in the DB is classified as a P-gp substrate or non-substrate (inconclusive or contradictive data are marked as Yes/No or No/Yes in the 'Substrate field) and efficiency of P-gp mediated transport is provided for substrates. High efficiency describes compounds that are transported with the rate similar to the best substrates (vinblastine, daunorubicin, paclitaxel). Similarly, drugs comprising the database are classified according to their Inhibitor liability. Potency field denotes effectivity of P-gp inhibition, High potency representing compounds that inhibit P-gp as good as standard inhibitor verapamil or even better.
In the Assays section, the methods that were used in the analysis of P-gp substrate/inhibitor specificity are listed:
- Substrate (in vitro transport assay) – polarized transport of drugs across P-gp expressing cell monolayers or decreased drug accumulation in MDR cells
- Substrate (in vivo BBB models) – increased distribution of drugs to the brain in P-gp deficient (mdr1a/b(-/-)) mice
- P-gp mediated resistance – P-gp overexpressing (MDR) cells demonstrate resistance to the drug
- Drug efflux inhibition – inhibition of drug efflux in P-gp expressing cells.
- MDR reversion – sensitization of P-gp expressing cells to “MDR profile” drugs (taxanes, anthracyclines, vinca alkaloids)
- P-gp ATPase modulation – activation or inhibition of P-gp ATPase. This assay does not differentiate P-gp substrates and inhibitors.
Model features & prediction accuracy
The model was developed with Algorithm Builder using a novel methodology consisting of two parts:
- Global baseline statistical model employing binomial PLS with multiple bootstrapping using a predefined set of fragmental descriptors.
- Local correction to baseline prediction based on analysis of experimental data for similar compounds.
The underlying methodology enables obtaining an intrinsic evaluation of prediction confidence by the means of Reliability Index (RI) values calculated for each prediction. RI ranging from 0 to 1 serves as an indication whether a submitted compound falls within the Model Applicability Domain. Two criteria influence the calculation of Reliability Index of a prediction:
- Similarity of the analyzed molecule to compounds in the Self-training Library (prediction is unreliable if no similar compounds have been found in the Library).
- Consistency of experimental data for similar compounds (discrepant data for similar molecules lead to lower RI values).
The presented method also forms the basis of model Trainability. 'Trainable model' methodology addresses the issue of the chemical space of ‘in-house’ libraries being considerably wider than that of publicly available data which results in limited applicability of most third-party QSARs for analysis of ‘in-house’ data. The ‘Training engine‘ makes appropriate corrections for systematic deviations produced by the baseline QSAR model based on analysis of similar compounds from the experimental data library. Expansion of this Self-training Library with user-defined experimental data for new compounds leads to instant improvement of prediction accuracy for the respective compound classes. Moreover, addition of 'in-house' data allows adapting the existing model to the particular experimental protocol used in your company and avoiding potential issues related to discrepancies between different experimental methods used for determination of drug interactions with P-gp.
If the compound is within model Applicability Domain (acceptable Reliability Index) accuracy and sensitivity of classification is close to 90% for both models.