P450 Substrates
Overview
Biotransformations of the molecule affect clearance rate of the drug, duration of the therapeutic effect, and formation of toxic metabolites that can have fatal consequence for the patient. Cytochrome P450 is the main family of enzymes responsible for xenobiotic metabolism in human organism. ACD/Percepta predictive models for cytochrome P450 Substrate specificity cover five major isoforms (CYP3A4, CYP2D6, CYP1A2, CYP2C9 and CYP2C19) accounting for absolute majority (>95%) of biotransformations mediated by cytochrome P450. These models have been developed using data sets ranging from 800 to 1000 compounds and provide the probability that compound of interest will be metabolized by a certain cytochrome P450 enzyme to a significant extent.
Features
- Calculates probability of the compound to be a substrate of 5 human cytochrome P450 isoforms (CYP3A4, CYP2D6, CYP1A2, CYP2C9 and CYP2C19)
- The quality of each prediction is evaluated quantitatively via the estimated Reliability Indices
- Predictions are visualized in the form of a bar plot
- Provides a list of up to five most similar structures from the training set with their experimental results and references
- Allows user to add experimental measurement data in order to expand the Applicability Domain of the Model
Interface
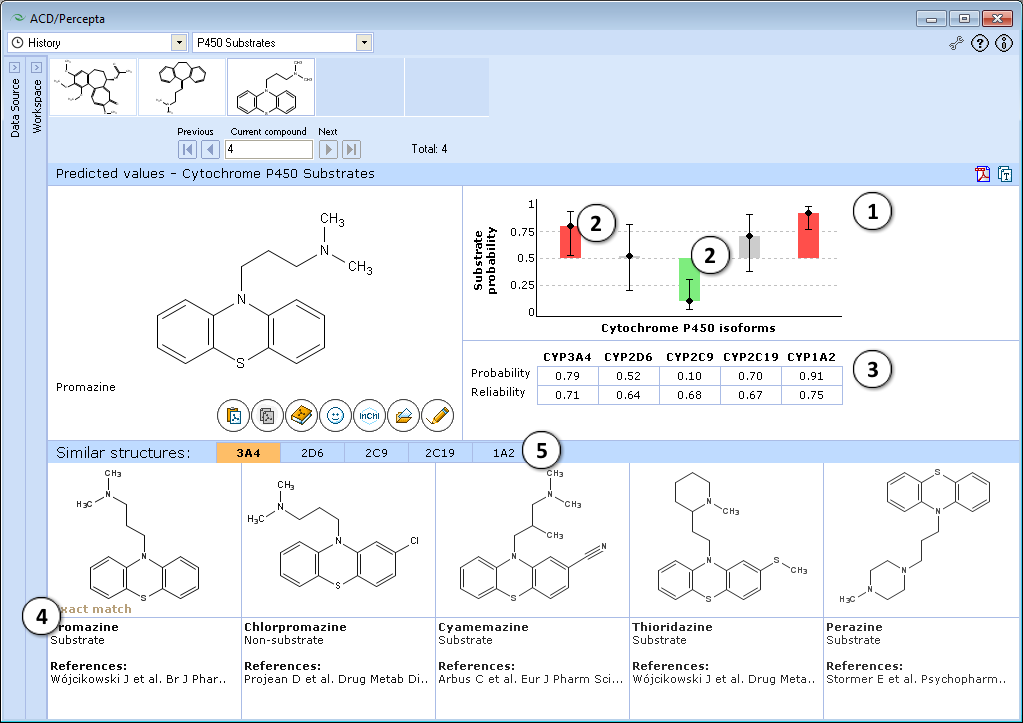
- Predictions are visualized in the form of bar charts.
- Height of the bar denotes estimated probability of being a substrate of the respective enzyme, whiskers indicate prediction intervals. Red bars represent confidently predicted substrates, green bars – confident non-substrates, and gray bars – inconclusive predictions. The coloring scheme takes into account both predicted probability and the Reliability Index values.
- The table below the chart shows exact values of calculated probabilities and Reliability Indices used to derive the bar plot.
- Five most similar structures from the training sets are displayed with experimental data and literature references.
- Click the corresponding tab to display similar structures for the relevant cytochrome P450 enzyme.
Technical information
Predicted endpoints
Cytochrome P450 Specificity-related modules in ACD/Percepta provide the following quantitative predictions:
- P450 Substrates: Probability that the compound of interest will be metabolized by a certain cytochrome P450 isoform.
- P450 Inhibitors: Probability that the compound of interest will inhibit a certain cytochrome P450 enzyme with IC50 below defined threshold. Two types of predictive models utilizing different IC50 thresholds have been developed. "General inhibition" models estimate whether the analyzed compound will exhibit any clinically significant cytochrome P450 inhibition at all (IC50 < 50 μM), while "Efficient inhibition" models predict probability that the compound will inhibit selected enzyme with IC50 < 10 μM.
- P450 Regioselectivity: Probability to be metabolized in human liver microsomes (or by a specific cytochrome P450 isoform) for every atom in the molecule.
Predictions are provided for five major cytochrome P450 isoforms (3A4, 2D6, 2C9, 2C19, 1A2) that are responsible for more than 80% of Phase I metabolism. In addition to Regioselectivity models for individual enzymes, overall HLM Regioselectivity module is also available. This module estimates the overall probabilities of human liver microsomal metabolism taking place at particular sites of the molecule. All predictions are supplied with Reliability Indices (RI) serving as an internal measure of prediction confidence (see Model Features section for more details about RI calculation).
Sources of experimental data
- P450 Metabolism (Substrate specificity and metabolism sites): only experimental data from original scientific publications were used for modeling of cytochrome P450 metabolism sites. The literature dataset was expanded with information about marketed drugs’ metabolism and the expanded dataset was used for cytochrome P450 substrate modeling. Two main types of assays were considered:
- Metabolism experiments using recombinant human cytochrome P450 enzymes
- Analysis of human liver microsomal (HLM) metabolism with contributions of a particular P450 isoform evaluated by evidence of metabolism inhibition with specific inhibitors of that isoform.
- P450 Inhibition: Inhibitor specificity data were also collected from original scientific publications based on HLM/recombinant enzyme assays, focusing on inhibition of metabolism of standard probe substrates by test compounds. Additionally, these databases were supplemented with information about marketed drugs, as well as HTS assay data from NCBI PubChem project.
Data sets
The sizes of the data sets used to develop the predictive models of substrate and inhibitor specificity are presented in the table below:
| Isoform | N (Substrate specificity) | N (Inhibitor specificity, cut-off: IC50 < 50 μM) | N (Efficient inhibition, cut-off: IC50 < 10 μM) |
|---|---|---|---|
| CYP1A2 | 935 | 4867 | 5815 |
| CYP2C9 | 867 | 7666 | 7677 |
| CYP2C19 | 794 | 6899 | 6833 |
| CYP2D6 | 1001 | 7707 | 7507 |
| CYP3A4 | 960 | 6684 | 7927 |
Regioselectivity models were based on experimental data for 873 compounds collected from publications dealing with analytical identification of the metabolites observed after the incubation of compound with human liver microsomes or recombinant cytochrome P450 enzymes. Every carbon atom with at least one hydrogen attached was marked as a site of metabolism, if hydroxylation at the atom was observed, or site of no metabolism otherwise. For dealkylation reactions, carbon atoms of the leaving groups were marked in the same manner. Some sites were marked as "inconclusive" and consequently not used in the modeling. The table below shows the overall number of marked atoms used for building the models:
| No. of atoms | HLM | CYP3A4 | CYP2D6 | CYP2C9 | CYP2C19 | CYP1A2 |
|---|---|---|---|---|---|---|
| Positive | 1269 | 795 | 354 | 288 | 249 | 383 |
| Inconclusive | 340 | 176 | 49 | 43 | 15 | 61 |
| Negative | 7182 | 6757 | 6305 | 6314 | 5210 | 6020 |
| Total | 8791 | 7728 | 6708 | 6645 | 5474 | 6464 |
Fully searchable Cytochrome P450 Specificity databases are not available in the current version of ACD/Percepta, yet each prediction performed by P450 Substrates and Inhibitors modules is displayed along with experimental data for five compounds from the training set most similar to the molecule of interest. The provided information for similar compounds includes classification as substrates/non-substrates (inhibitors/non-inhibitors at two IC50 cut-offs) of the relevant cytochrome P450 enzyme assigned on the basis of experimental results together with original references. In case of Regioselectivity predictions five most similar atoms from the training set are shown with color-marks indicating whether a metabolic reaction taking place at the particular site of the molecules was observed experimentally.
Model features & prediction accuracy
The predictive models of all cytochrome P450-related endpoints were derived using GALAS (Global, Adjusted Locally According to Similarity) modeling methodology (please refer to [1] for more details).
Each GALAS model consists of two parts:
- Global baseline statistical model employing binomial PLS with multiple bootstrapping using a predefined set of fragmental descriptors, that reflects general trends in the considered property.
- Similarity-based routine that performs local correction of baseline predictions taking into account the differences between baseline and experimental values for the most similar training set compounds.
GALAS methodology also provides the basis for estimating reliability of predictions by the means of calculated Reliability Index (RI) value that takes into account:
- Similarity of tested compound to the training set molecules (prediction is unreliable if no similar compounds have been found).
- Consistence of experimental values and baseline model prediction for the most similar similar compounds from the training set (discrepant data for similar molecules lead to lower RI values).
Reliability Index ranges from 0 to 1 (0 corresponds to a completely unreliable, and 1 - a highly reliable prediction) and serves as an indication whether a submitted compound falls within the Model Applicability Domain. Compounds obtaining predictions RI < 0.3 are considered outside of the Applicability Domain of the model.
The predictive models of cytochrome P450 substrate and inhibitor specificity are also Trainable meaning that their Applicability Domains may be expanded to account for the ‘in-house’ experimental data available in your company without the need to rebuild the baseline statistical model from scratch. Addition of new compounds to the module Self-training Library results in an instant improvement of prediction accuracy for the respective compound classes. Moreover, addition of 'in-house' data allows adapting the existing model to the particular experimental protocol used in your company and avoiding potential issues related to discrepancies between different experimental methods used for determination of drug interactions with CYP450 enzymes.
If the compound is within model Applicability Domain (acceptable Reliability Index) accuracy and sensitivity of classification is close to 90% for inhibitors and close to 80% for substrates. The accuracy of in silico prediction of cytochrome P450 inhibition is comparable to the screening results.
The results of the external validation of CYP3A4 Inhibition prediction model and a demonstration of model training can be found in an Application Note [2]. For information on the competitive performance of HLM Regioselectivity model please refer to [3].
Publications
More detailed information about the construction of databases, and application of GALAS method to modeling cytochrome P450-related properties can be found in the following articles: