Trainable Models
The ‘Trainable Model’ concept utilizing a novel similarity based analysis methodology allows the user to:
- Assess the quality of the predictions by means of the Reliability Index (RI) estimation. This index provides values in a range from 0 to 1 and serves as an evaluation of whether a submitted compound falls within the Model Applicability Domain. Estimation of the Reliability Index takes into account the following two aspects: similarity of the tested compound to the training set and the consistency of experimental values for similar compounds.
- Instantly expand the Model Applicability Domain with the help of any user-defined proprietary ‘in-house’ data of experimental values for the property of interest.
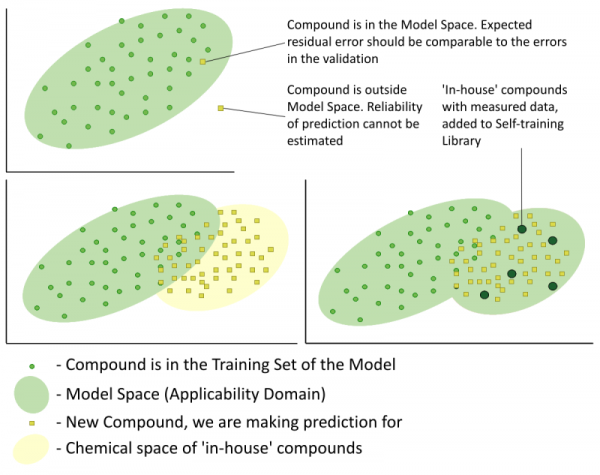
Each ‘Trainable Model’ consists of the following parts:
- A structure based QSAR/QSPR for the prediction of the property of interest derived from a literature training set – the baseline QSAR/QSPR.
- A user defined data set with experimental values for the property of interest – the Self-training Library.
- A special similarity based routine which identifies the most similar compounds contained in the Self-training Library and considering their experimental values calculates systematic deviations produced by the baseline QSAR/QSPR for each submitted molecule – the training engine.
The current version of ACD/Percepta has implemented ‘Trainable Model’ methodology for the prediction of the following properties:
- P-gp Specificity
- Trainable P-gpS
Calculates the probability of a compound being a P-gp substrate. - Trainable P-gpI
Predicts the probability for a compound to act as a P-gp inhibitor.
- Trainable P-gpS
- Solubility
- Trainable LogS0
Calculates intrinsic solubility in water (LogS0, mmol/ml). - Trainable LogS
Calculates solubility in buffer at relevant pH values (LogS, mmol/ml).
- Trainable LogS0
- Plasma Protein Binding
- Trainable LogKa(HSA)
Predicts the compound's equilibrium binding constant to human serum albumin in the blood plasma (LogKaHSA). - Trainable PPB
Estimates the fraction of the compound bound to the blood plasma proteins (%PPB)
- Trainable LogKa(HSA)
- Partitioning
- Trainable LogP
Calculates the logarithm of the octanol-water partitioning coefficient for the neutral form of the compound (LogP) - Trainable LogD
Calculates the logarithm of the apparent octanol water partition coefficient at relevant pH values (LogD) taking into account all the species (including ionized) of the compound present in the solution.
- Trainable LogP
- Cytochrome P450 Inhibitor Specificity
Calculates probability of a compound being an inhibitor of a particular cytochrome P450 enzyme with IC50 below one of the two selected thresholds (general inhibition models - IC50 < 50 μM; efficient inhibition - IC50 < 10 μM). Predictions are available for five P450 isoforms :- Trainable CYP1A2 I
- Trainable CYP2C19 I
- Trainable CYP2C9 I
- Trainable CYP2D6 I
- Trainable CYP3A4 I
- Cytochrome P450 Substrate Specificity Calculates probability of a compound being metabolized by a particular cytochrome P450 enzyme. Predictions are available for five P450 isoforms:
- Trainable CYP1A2 S
- Trainable CYP2C19 S
- Trainable CYP2C9 S
- Trainable CYP2D6 S
- Trainable CYP3A4 S
As a starting point for the calculations a number of Built-in Self-training Libraries with experimental values of the corresponding properties is provided for each ‘Trainable Model’ in ACD/Percepta.
For more information see Trainable Libraries and Training