LogD: Difference between revisions
Jump to navigation
Jump to search
| Line 13: | Line 13: | ||
# Displays several logD values at physiologically relevant pHs. [[File:Logd_editph.png|right]]<br>a. Click "+/-" nodes to expand/collapse the option of new pH point's addition.<br>b. Enter desirable pH value and press "Add". The new pH point along with automatically calculated logD value will appear at the end of the list.<br>c. Hover over the pH point and press the "x" pictogram to remove it from the list. | # Displays several logD values at physiologically relevant pHs. [[File:Logd_editph.png|right]]<br>a. Click "+/-" nodes to expand/collapse the option of new pH point's addition.<br>b. Enter desirable pH value and press "Add". The new pH point along with automatically calculated logD value will appear at the end of the list.<br>c. Hover over the pH point and press the "x" pictogram to remove it from the list. | ||
# Press "Configure" button to select LogP and pKa calculation algorithms, and libraries to use for training, or set other options. | # Press "Configure" button to select LogP and pKa calculation algorithms, and libraries to use for training, or set other options.<br>'''NOTE:''' Training takes place through LogP Self-training Libraries. The training procedure implemented in ACD/LogP GALAS module accepts LogD values measured at any pH and automatically recalculates these to the respective LogP of neutral species to be stored in the library. The trained LogP library may then be used in both LogP and LogD calculations. | ||
# Click and drag the slider to see calculated logD at precise pH value displayed on the right. | # Click and drag the slider to see calculated logD at precise pH value displayed on the right. | ||
<br /> | <br /> | ||
Revision as of 13:55, 21 January 2013
Overview
This module calculates the logarithm of the apparent octanol-water partition coefficient D for all (including ionized) compound species at various pH values. Prediction results are presented in two ways – an easy to navigate and interpret graph and a list of LogD predictions at discrete pH values. Initially this list is populated by the LogD predictions at physiologically relevant pH conditions; however it is fully customizable via the ability to add or remove the predictions at any pH value.
Interface
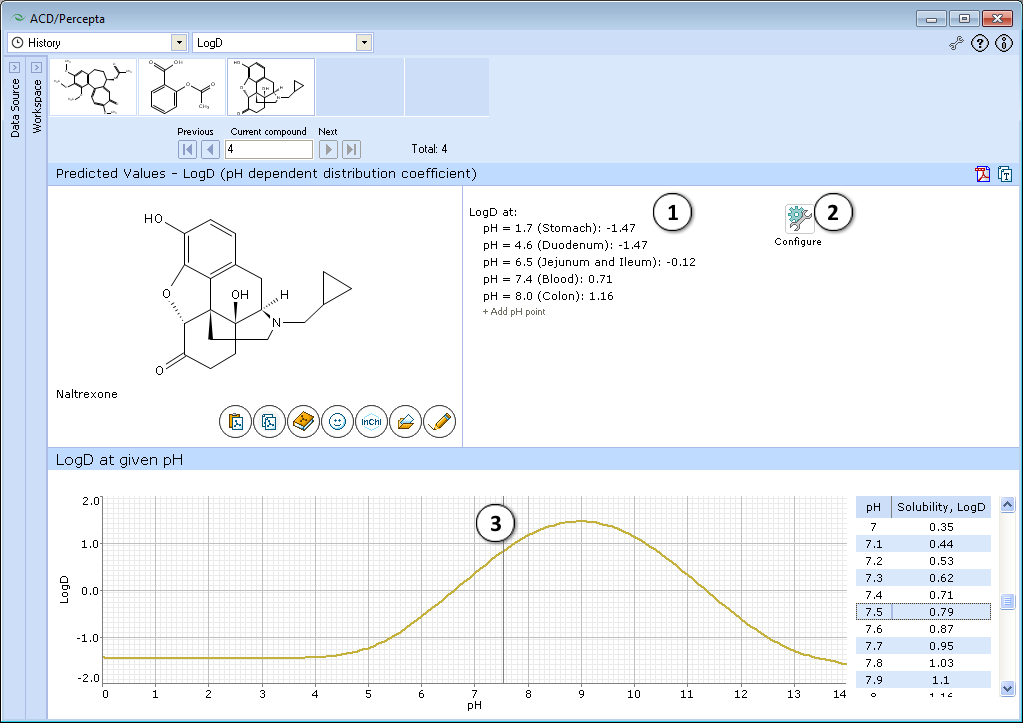
- Displays several logD values at physiologically relevant pHs.

a. Click "+/-" nodes to expand/collapse the option of new pH point's addition.
b. Enter desirable pH value and press "Add". The new pH point along with automatically calculated logD value will appear at the end of the list.
c. Hover over the pH point and press the "x" pictogram to remove it from the list. - Press "Configure" button to select LogP and pKa calculation algorithms, and libraries to use for training, or set other options.
NOTE: Training takes place through LogP Self-training Libraries. The training procedure implemented in ACD/LogP GALAS module accepts LogD values measured at any pH and automatically recalculates these to the respective LogP of neutral species to be stored in the library. The trained LogP library may then be used in both LogP and LogD calculations. - Click and drag the slider to see calculated logD at precise pH value displayed on the right.