Fragmentate Menu: Difference between revisions
Jump to navigation
Jump to search
Created page with "File:spreadsheet_fragmentate_menu.png <br> <br> <ol> <li>'''Retrosynthetic''' – runs Retrosynthetic fragmentation. Fragments are obtained by cleaving bonds that can be o..." |
No edit summary |
||
| Line 3: | Line 3: | ||
<br> | <br> | ||
<ol> | <ol> | ||
<li>'''Retrosynthetic''' – runs Retrosynthetic fragmentation. Fragments are obtained by cleaving bonds that can be obtained with the help of some popular reactions of organic synthesis, e. g. esterification. In the dialog box that opens first you can select which fields of the active spreadsheet will be transferred to the fragment spreadsheet (this helps to relate fragments and initial molecules). The newly created dataset appears in the Data Source | <li>'''Retrosynthetic''' – runs Retrosynthetic fragmentation. Fragments are obtained by cleaving bonds that can be obtained with the help of some popular reactions of organic synthesis, e. g. esterification. In the dialog box that opens first you can select which fields of the active spreadsheet will be transferred to the fragment spreadsheet (this helps to relate fragments and initial molecules). The newly created dataset appears in the Data Source panel Substituents group under automatic name ''Retrosynth''.</li><br> | ||
<li>'''Find and Replace Fragments''' – runs “Find and Replace” fragmentation. It replaces certain functional groups by R-symbols. You will need a reaction database file (*.RDB) that contains the rules of fragmentation. In the dialog box that opens you can select the reaction database and the fields you want to transfer to the fragment spreadsheet:<br><br> | <li>'''Find and Replace Fragments''' – runs “Find and Replace” fragmentation. It replaces certain functional groups by R-symbols. You will need a reaction database file (*.RDB) that contains the rules of fragmentation. In the dialog box that opens you can select the reaction database and the fields you want to transfer to the fragment spreadsheet:<br><br> | ||
[[File:spreadsheet_fragmentate_menu_findandreplace.png]]<br><br> | [[File:spreadsheet_fragmentate_menu_findandreplace.png|center]]<br><br> | ||
<ol style="list-style-type:lower-latin"> | <ol style="list-style-type:lower-latin"> | ||
<li>Select the reaction database from the dropdown list or browsing your local PC drive only if it was created. To create it, click the '''Reaction Editor''' button (see below).</li> | <li>Select the reaction database from the dropdown list or browsing your local PC drive only if it was created. To create it, click the '''Reaction Editor''' button (see below).</li> | ||
| Line 13: | Line 13: | ||
</ol> | </ol> | ||
</li> | </li> | ||
The newly created dataset with reactions appears in the | The newly created dataset with reactions appears in the Data Source panel Substituents group under the automatic name ''ReplacedFragments''.</li> | ||
<br> | <br> | ||
<li>'''Reaction Editor''' – opens the '''Reaction Editor''' window to create or modify the set of reactions (*.RDB file). </li> | <li>'''Reaction Editor''' – opens the '''Reaction Editor''' window to create or modify the set of reactions (*.RDB file). </li> | ||
<br> | <br> | ||
[[File:spreadsheet_fragmentate_menu_reactioneditor1.png]] | [[File:spreadsheet_fragmentate_menu_reactioneditor1.png|center]] | ||
<br><br> | <br><br> | ||
<ol style="list-style-type:lower-latin"> | <ol style="list-style-type:lower-latin"> | ||
| Line 27: | Line 27: | ||
<li>Use + or – to add/remove one more reaction.</li> | <li>Use + or – to add/remove one more reaction.</li> | ||
<br> | <br> | ||
[[File:spreadsheet_fragmentate_menu_reactioneditor2.png|1000px]] | [[File:spreadsheet_fragmentate_menu_reactioneditor2.png|center|1000px]] | ||
<br> | <br> | ||
<br> | <br> | ||
Revision as of 14:35, 4 July 2012
- Retrosynthetic – runs Retrosynthetic fragmentation. Fragments are obtained by cleaving bonds that can be obtained with the help of some popular reactions of organic synthesis, e. g. esterification. In the dialog box that opens first you can select which fields of the active spreadsheet will be transferred to the fragment spreadsheet (this helps to relate fragments and initial molecules). The newly created dataset appears in the Data Source panel Substituents group under automatic name Retrosynth.
- Find and Replace Fragments – runs “Find and Replace” fragmentation. It replaces certain functional groups by R-symbols. You will need a reaction database file (*.RDB) that contains the rules of fragmentation. In the dialog box that opens you can select the reaction database and the fields you want to transfer to the fragment spreadsheet:
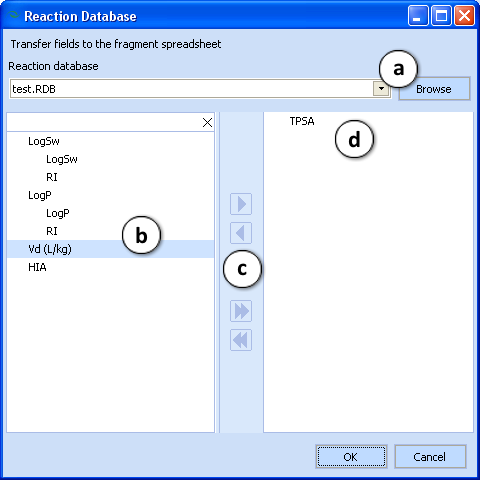
- Select the reaction database from the dropdown list or browsing your local PC drive only if it was created. To create it, click the Reaction Editor button (see below).
- Select which fields of the active spreadsheet will be transferred to the fragment spreadsheet.
- Move one by one or all fields at once between the panes.
- The selected fields.
The newly created dataset with reactions appears in the Data Source panel Substituents group under the automatic name ReplacedFragments.
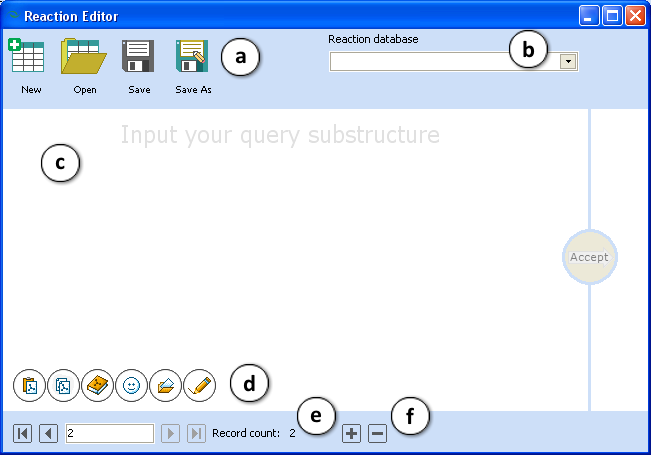
- Reaction Editor – opens the Reaction Editor window to create or modify the set of reactions (*.RDB file).
- Standard File menu commands to create new, open the existing or save the current database of reactions.
- Select the previously created reaction database from the dropdown list if needed.
- The Structure pane
- Standard commands to get a 2-D input structure
- Status line indicating the current number of visible reaction and the total number of records in the database.
- Use + or – to add/remove one more reaction.
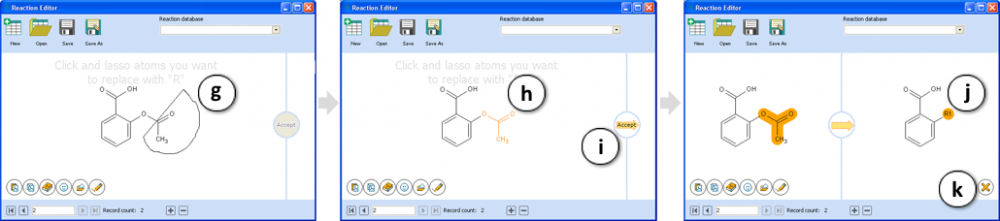
- After the input structure is loaded, click and lasso the atoms you want to replace with “R” (substituent).
- The selection is highlighted in orange. If you wish to change selection, re-lasso the atoms.
- Click Accept to confirm selection.
- The marked structure fragment is replaced with R1.
- Click to cancel the created reaction.