Active Transport: Difference between revisions
No edit summary |
|||
| Line 14: | Line 14: | ||
<br /> | <br /> | ||
[[Image: | [[Image:Active_transport.png|center]] | ||
<br /> | <br /> | ||
Revision as of 10:17, 8 July 2013
Overview
Active transport module offers classification methods for the prediction of the compound transport across intestinal membrane by oligopeptide transporter (PepT1) or bile acid transporter (ASBT) as well as the similarity based assessment of the likelihood of other most important active transport systems (monocarboxylic acid transporter (MCT1), amino acid carriers) involvement in compound's ability to penetrate intestinal barrier.
Features
- PepT1 (oligopeptide transporter) and ASBT (bile acid transporter) substrate prediction
- Similarity approach used for prediction of other possible active transport systems (monocarboxylic acid transporter
- MCT1, amino acid carriers, etc.)
- Provides reference data for similar structures
Interface
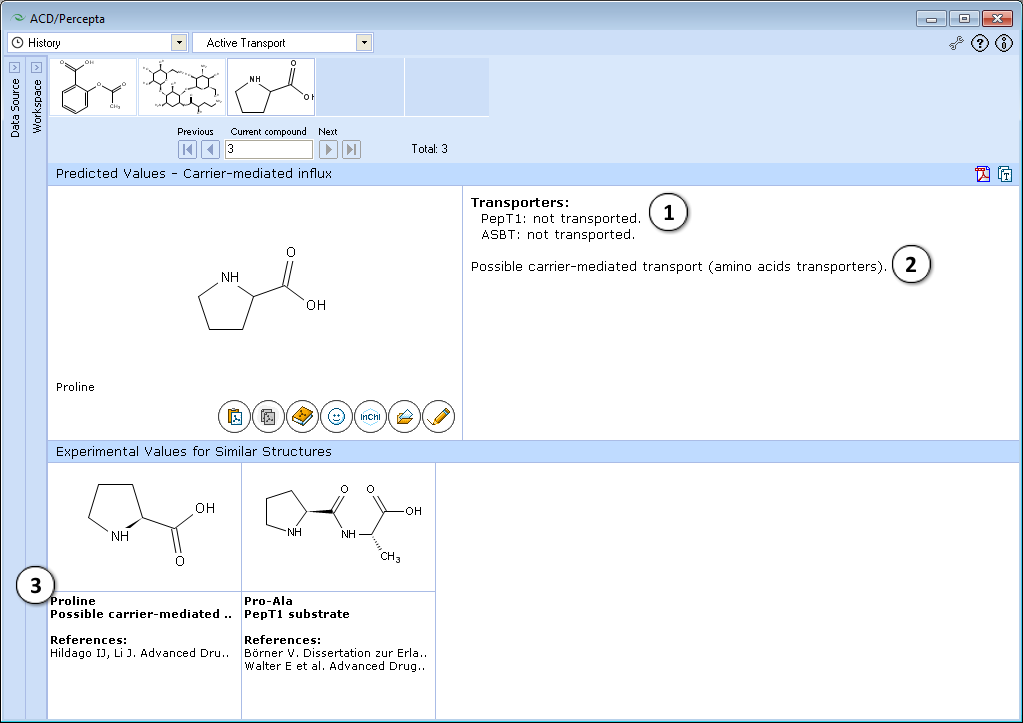
- Prediction of PepT1 and ASBT transport
- Prediction for other transport systems (MCT1, amino acid carriers, etc.)
- Up to 5 most similar structures in the training set with experimental values and references
Technical information
Training set size:
| Model | Number of compounds |
| PepT1 | N/A* |
| ASBT | N/A* |
| Other transporters | N/A* |
* - All ACD/Active Transport models are knowledge based "expert-like" systems, therefore the concept of training set does not apply to the process of their development.
Validation set size:
| Model | Number of compounds |
| PepT1 | 330 |
| ASBT | 173 |
| Other transporters | N/A* |
* - The substrate specificity of the remaining transporters is very narrow. The compounds that can be transported by these enzymes are considered to be well known and documented. As a result any actual experimental measurements for new compounds are very rare and no adequate validation sets could be compiled to test the accuracy of the classification models in question.
Main sources of experimental data:
- Various articles from peer-reviewed scientific journals*
* - Both articles reporting corresponding Active Transport models by other authors (i.e. with larger collections of experimental data per article) and dealing with detailed experimental pharmacokinetic characterization (i.e. usually only several compounds per article) were available for the validation set construction.
Internal Validation
| Subset | Coverage of the entire internal validation set (N=330) |
Observed | Assigned class | |||
|---|---|---|---|---|---|---|
| Transported | Not-transported | |||||
| Compounds receiving definite model predictions N = 311* |
|
True | 121 (38.9%) |
10 (3.2%) | ||
| False | 13 (4.2%) |
167 (53.7%) | ||||
| Accuracy |
| |||||
| Sensitivity |
| |||||
| Specificity |
| |||||
* - 'No predcition' is one of the possible outputs of the classification model, meaning that it cannot make an unambiguous judgment based on available rules. Obviously such compounds cannot be included in the accuracy assessment. As a result not all compounds fall into the applicability domain of the model even with no additional parameters (i.e., RI) available to filter out unreliable predictions as in GALAS models. Analysis of experimental data for similar compounds can still be used as a source of information regarding active influx via PepT1 transporter for the compounds which cannot be classified by the model.
| Subset | Observed | Assigned class | |||
|---|---|---|---|---|---|
| Transported | Not-transported | ||||
| Entire internal validation set N = 173* |
True | 22 (12.7%) |
8 (4.6%) | ||
| False | 7 (4.0%) |
136 (78.6%) | |||
| Accuracy |
| ||||
| Sensitivity |
| ||||
| Specificity |
| ||||
* - There are no inconclusive predictions produced by the model. Moreover, since ACD/Active Transport models do not utilize GALAS modeling methodology, no additional parameters (i.e., RI) are available to enable additional filtering of more reliable predictions.