Vd: Difference between revisions
Created page with "==Overview== <br /> This module contains a predictive algorithm that generates a quantitative estimate of the apparent volume of distribution of a compound. Physicochemical p..." |
|||
| Line 2: | Line 2: | ||
<br /> | <br /> | ||
This module contains a predictive algorithm that generates a quantitative estimate of the apparent | This module contains a predictive algorithm that generates a quantitative estimate of the apparent Volume of Distribution (Vd) of a compound. Physicochemical parameters, charge state, lipophilicity and hydrogen bonding capacity are automatically computed and used as inputs to the predictive model of the volume of distribution.<br /> | ||
===Features=== | ===Features=== | ||
* Calculates the extent of tissue binding of candidate compounds expressed as Vd values and displays a comment regarding the effect of physicochemical properties (LogP and ionization) on drug distribution in the body. | |||
* Experimentally measured volume of distribution is displayed for up to 5 similar structures from the training set along with the corresponding literature references. | |||
* | |||
* Experimentally measured | |||
* A clear and straightforward interface ensures quick and easy addition of user-defined data to the Self-training Library. | * A clear and straightforward interface ensures quick and easy addition of user-defined data to the Self-training Library. | ||
<br /> | <br /> | ||
Revision as of 13:51, 17 May 2012
Overview
This module contains a predictive algorithm that generates a quantitative estimate of the apparent Volume of Distribution (Vd) of a compound. Physicochemical parameters, charge state, lipophilicity and hydrogen bonding capacity are automatically computed and used as inputs to the predictive model of the volume of distribution.
Features
- Calculates the extent of tissue binding of candidate compounds expressed as Vd values and displays a comment regarding the effect of physicochemical properties (LogP and ionization) on drug distribution in the body.
- Experimentally measured volume of distribution is displayed for up to 5 similar structures from the training set along with the corresponding literature references.
- A clear and straightforward interface ensures quick and easy addition of user-defined data to the Self-training Library.
Interface
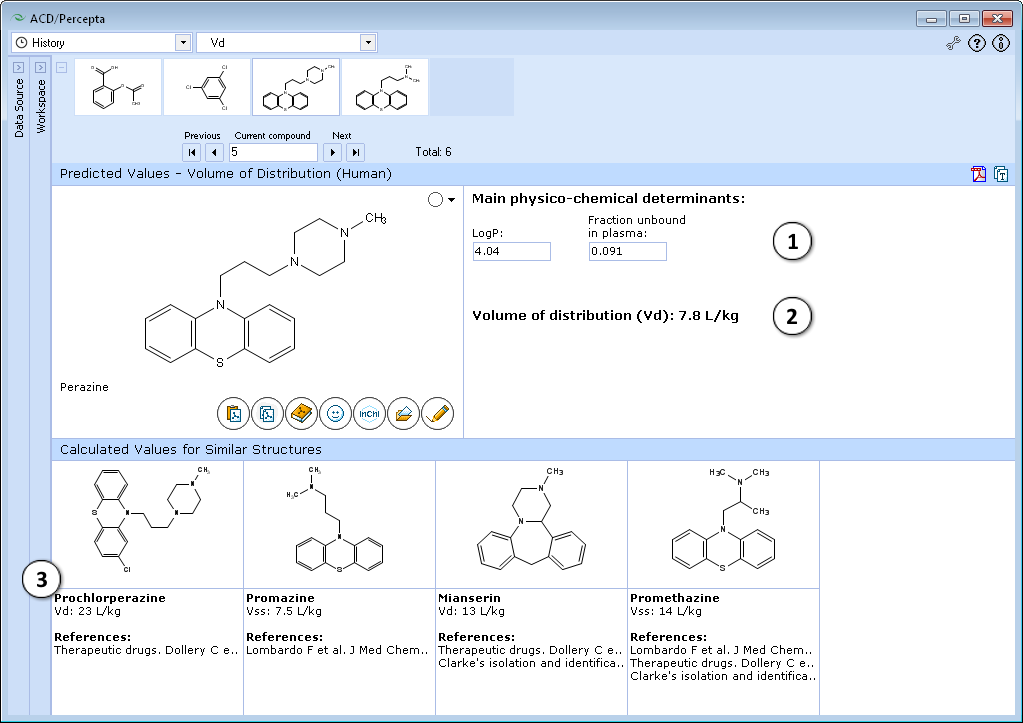
- Calculated apparent volume of distribution of a compound:
File:distribution vd scale.png - A general estimate of likely volume of distribution for compounds of this type, based on chemical structure and physicochemical properties
- Up to 5 similar structures in the training set with experimental quantitative values of apparent volume of distribution and literature references
Technical information
Calculated quantitative parameters
Parameters calculated by Distribution\Protein Binding module include percentage plasma protein binding values (%PPB) and log KaHSA constants. These properties are related, but characterize provide slightly different information about the considered process.
- log KaHSA represents the drug’s affinity constant to human serum albumin – the major carrier protein in plasma. Experimental data come from direct chromatographic determination of binding strength to that particular protein.
log KaHSA = log ([LA]/([L][A])) where [LA] is concentration of ligand bound to albumin, [L] – that of free ligand, and [A] – concentration of free albumin which is estimated at ~0.6 mM in human plasma.
- %PPB values represent the overall fraction of drug bound in human plasma, i.e. accounts for interactions with different proteins: albumin, alpha1-acid glycoprotein, lipoproteins, SHBG, transcortin etc. In vitro measurements of the extent of plasma protein binding usually involve equilibrium dialysis, ultrafiltration or ultracentrifugation methods.
%PPB = (1 – fu) * 100% where fu is fraction of free (unbound) drug in plasma ranging from 0 to 1.
- Supplementary Distribution\Vd module calculates apparent Volume of Distribution of drugs in human body expressed in liters per kg body weight (L/kg).
Experimental data
Experimental data that were utilized to build predictive models were collected from drug prescription information, reference pharmacokinetic tabulations and many original articles. The main sources of Vd data were well-known pharmacokinetic books: "Therapeutic Drugs" (ed. by C. dollery), and Goodman & Gilman's "The Pharmacological Basis of Therapeutics", while albumin affinity constants were collected mainly from original articles by Valko K. et al. J Pharm Sci. 2003;92(11):2236-48. [1], and Kratochwil N.A. et al. Biochem Pharmacol. 2002;64(9):1355-74. [2] The compiled data sets contain %PPB data for almost 1500 compounds, about 340 albumin affinity constants and almost 800 Vd values.
Model development (technical details)
The models for predicting %PPB and log KaHSA constants were developed with Algorithm Builder using a novel methodology consisting of two parts:
- Global baseline statistical model employing binomial PLS with multiple bootstrapping using a predefined set of fragmental descriptors.
- Local correction to baseline prediction based on analysis of experimental data for similar compounds.
The underlying methodology enables obtaining an intrinsic evaluation of prediction confidence by the means of Reliability Index (RI) values calculated for each prediction. RI ranging from 0 to 1 serves as an indication whether a submitted compound falls within the Model Applicability Domain. Two criteria influence the calculation od Reliability Index of a prediction:
- Similarity of the analyzed molecule to compounds in the Self-training Library (prediction is unreliable if no similar compounds have been found in the Library).
- Consistency of experimental data for similar compounds – reliability of calculated values is lower is data for similar compounds are discrepant.
Both %PPB and log KaHSA predictive models are Trainable meaning that their Applicability Domains may be easily extended by addition of ‘in-house’ experimental data to the module Self-training Library. Notably, the baseline statistical model does not need to be rebuilt from scratch to account for data entered by the user. The model is retrained automatically as new compounds are added to the Library. Model trainability would be particularly useful for predicting serum albumin affinity constants as literature data sets are very sparse, thus the ability to take advantage of large ‘in-house’ libraries gives the potential for a significant improvement of both accuracy and reliability of calculations.