Aquatic Toxicity LC50: Difference between revisions
Updated for 2024 release |
|||
| (5 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<br /> | <br /> | ||
Aquatic toxicity module provides the researcher with an accurate and reliable predictive tool that may serve as a valuable first estimate of fish and | Aquatic toxicity module provides the researcher with an accurate and reliable predictive tool that may serve as a valuable first estimate of fish, daphnid, and protozoan toxicity of new chemical entities that is required under REACH. It may therefore be used as an initial screen that could compete and become at least a partial replacement of time and resource consuming experimental determination in animals.<br /> | ||
===Features=== | ===Features=== | ||
* | * Calculates short-term toxicity to three species that are typically used in aquatic toxicity assays. The predicted parameters include lethal concentration (LC50, mg/L) to fathead minnows (''Pimephales promelas'') and water fleas (''Daphnia magna''), as well as inhibitory growth concentration (IGC50, mg/L) to ciliate protozoa (''Tetrahymena pyriformis''). | ||
* The calculated toxicity values are supported by reliability indices (RI) that provide a quantitative evaluation of prediction confidence. High RI shows that the calculated value is likely to be accurate, while low RI indicates that no similar compounds with consistent data are present in the training set. | |||
* The calculated | * Displays 5 most similar compounds from the training set for each considered aquatic species along with experimental toxicity values and similarities to the currently analyzed compound. | ||
* The training sets used to build the models contain experimental data on aquatic toxicity to fathead minnows for about 900 compounds, to water fleas for about 600 compounds, and to ciliate protozoa for about 1100 compounds.<br /> | |||
* The training sets used to build the models contain experimental data on aquatic toxicity for about 900 compounds | |||
<br /> | <br /> | ||
<span style="color:red; font-weight: bold;">IMPORTANT NOTE:</span> | |||
If you installed Percepta as an upgrade over a previous version, the program will attempt to preserve any custom configuration of Self-training libraries used in Aquatic Toxicity module. This configuration will not include the new, significantly extended built-in libraries that were introduced in 2024 release. In this case, to take advantage of the new libraries, you may need to click "Configure" for the respective endpoint and manually select the following entries: ''LC50 (D. magna) v. 1.3 (read-only)'', ''IGC50 (T. pyriformis) v. 1.1 (read-only)''. | |||
In case of a new installation, the new library should be selected automatically with no further action required. | |||
== Interface == | == Interface == | ||
| Line 19: | Line 24: | ||
<br /> | <br /> | ||
# Calculations are presented in the form of a table. Each row contains dedicated "Configure" and "Train" buttons to select the training library for the particular species and to add new data to that library. Predictions are made for the | # Calculations are presented in the form of a table. Each row contains dedicated "Configure" and "Train" buttons to select the training library for the particular species and to add new data to that library, as well as "QPRF" button to generate the QPRF report in PDF format. Predictions are made for the three aquatic species most frequently used for testing - fathead minnows (''Pimephales promelas''), water fleas (''Daphnia magna''), and ciliate protozoa (''Tetrahymena pyriformis''). | ||
# The predicted value is LC50 of the analyzed compound | # The predicted value is either LC50, or IGC50 of the analyzed compound to a given organism, expressed in mg/L. | ||
# Predictions are supported by Reliability Index values ranging from 0 to 1 that serve as an intrinsic evaluation of prediction confidence: | # Predictions are supported by Reliability Index values ranging from 0 to 1 that serve as an intrinsic evaluation of prediction confidence: | ||
#* RI < 0.3 – Not Reliable, | #* RI < 0.3 – Not Reliable, | ||
| Line 26: | Line 31: | ||
#* RI in range 0.5-0.75 – Moderate Reliability, | #* RI in range 0.5-0.75 – Moderate Reliability, | ||
#* RI >= 0.75 – High Reliability | #* RI >= 0.75 – High Reliability | ||
# Up to five most similar compounds from the training set with names, CAS numbers and experimental LC50 values. | # Up to five most similar compounds from the training set with names, CAS numbers and experimental LC50/IGC50 values. | ||
# Click the tab to browse the similar structures for different species. | # Click the tab to browse the similar structures for different species. | ||
<br /> | <br /> | ||
<div class="mw-collapsible | <div class="mw-collapsible"> | ||
==Technical information== | ==Technical information== | ||
| Line 40: | Line 45: | ||
<br /> | <br /> | ||
The standard measure of aquatic toxicity is the concentration of the compound in water that is lethal to 50% of exposed organisms (LC50). In the case of protozoa, the endpoint is not lethality, but inhibition of cell growth by 50% (IGC50). To obtain a linear relationship with structural properties these data were converted to logarithmic form (pLC50/pIGC50) for modeling, but the final prediction result is returned as an original LC50/IGC50 value in mg/L. | |||
===Experimental data=== | ===Experimental data=== | ||
Experimental data | Experimental data used for the development of predictive models were collected from EPA reference databases, as well as original publications. | ||
After thorough verification of the obtained values the final data sets contained about 900 compounds with quantitative LC50 values characterizing acute toxicity to fishes (''Pimephales promelas''), | After thorough verification of the obtained values the final data sets contained about 900 compounds with quantitative LC50 values characterizing acute toxicity to fishes (''Pimephales promelas''), 600 compounds - to water fleas (''Daphnia magna''), and 1100 compounds - to ciliate protozoa (''Tetrahymena pyriformis''). | ||
===Model features & prediction accuracy=== | ===Model features & prediction accuracy=== | ||
The predictive models | The predictive models were derived using GALAS (Global, Adjusted Locally According to Similarity) modeling methodology (please refer to [http://www.ncbi.nlm.nih.gov/pubmed/20373217] for more details). | ||
Each GALAS model consists of two parts: | Each GALAS model consists of two parts: | ||
| Line 64: | Line 69: | ||
The resulting models are highly accurate: LC50 values for aquatic species are predicted with RMSE of 0.5-0.6 log units when only predictions of moderate and high reliability (''RI'' >= 0.5) are considered .''RI'' values in the high and moderate ranges are commonly obtained for 30-60% of the validation sets. Validation results also show that the accuracy of predictions is proportional to the Reliability Index, as shown in the table below for LC50 to fishes (''P. promelas''): | The resulting models are highly accurate: LC50 values for aquatic species are predicted with RMSE of 0.5-0.6 log units when only predictions of moderate and high reliability (''RI'' >= 0.5) are considered .''RI'' values in the high and moderate ranges are commonly obtained for 30-60% of the validation sets. Validation results also show that the accuracy of predictions is proportional to the Reliability Index, as shown in the table below for LC50 to fishes (''P. promelas''): | ||
{| cellpadding="2" cellspacing="0" style="border-top: | {| cellpadding="2" cellspacing="0" style="border-top:1px solid black; border-bottom:1px solid black" | ||
|- | |- | ||
! style="border-bottom:1px; background:#EAEAEA" width="150" | Subset | ! style="border-bottom:1px; background:#EAEAEA" width="150" | Subset | ||
| Line 72: | Line 76: | ||
! style="border-bottom:1px; background:#EAEAEA" width="100" | <i>RMSE</i> | ! style="border-bottom:1px; background:#EAEAEA" width="100" | <i>RMSE</i> | ||
|- | |- | ||
| align="center | | align="center" | ''RI'' > 0.3 | ||
| align="center" | | | align="center" | | ||
{| cellpadding=" | {| cellpadding="2" cellspacing="0" style="width:80%; height:40px" | ||
| style=" | | style="background:#B9CDE5" align="right" width="85.7%" | '''85.7%''' || style="background:#EDF2F9" width="14.3%" | | ||
|} | |} | ||
| align="center" | 0.656 || align="center" | 0.797 | | align="center" | 0.656 || align="center" | 0.797 | ||
|- | |- | ||
| align="center | | align="center" | ''RI'' > 0.5 | ||
| align="center" | | | align="center" | | ||
{| cellpadding=" | {| cellpadding="2" cellspacing="0" style="width:80%; height:40px" | ||
| style=" | | style="background:#B9CDE5" align="right" width="59.4%" | '''59.4%''' || style="background:#EDF2F9" width="41.6%" | | ||
|} | |} | ||
| align="center" | 0.795 || align="center" | 0.501 | | align="center" | 0.795 || align="center" | 0.501 | ||
|- | |- | ||
| align="center | | align="center" | ''RI'' > 0.7 | ||
| align="center" | | | align="center" | | ||
{| cellpadding=" | {| cellpadding="2" cellspacing="0" style="width:80%; height:40px" | ||
| style=" | | style="background:#B9CDE5" align="right" width="28.0%" | '''28.0%''' || style="background:#EDF2F9" width="72.0%" | | ||
|} | |} | ||
| Line 100: | Line 104: | ||
|} | |} | ||
For more information regarding the modeling principles and validation results | For more information regarding the modeling principles and validation results please refer to [http://perceptahelp.acdlabs.com/docs/Aquatic_Tox.pdf]. | ||
Latest revision as of 08:11, 24 September 2024
Overview
Aquatic toxicity module provides the researcher with an accurate and reliable predictive tool that may serve as a valuable first estimate of fish, daphnid, and protozoan toxicity of new chemical entities that is required under REACH. It may therefore be used as an initial screen that could compete and become at least a partial replacement of time and resource consuming experimental determination in animals.
Features
- Calculates short-term toxicity to three species that are typically used in aquatic toxicity assays. The predicted parameters include lethal concentration (LC50, mg/L) to fathead minnows (Pimephales promelas) and water fleas (Daphnia magna), as well as inhibitory growth concentration (IGC50, mg/L) to ciliate protozoa (Tetrahymena pyriformis).
- The calculated toxicity values are supported by reliability indices (RI) that provide a quantitative evaluation of prediction confidence. High RI shows that the calculated value is likely to be accurate, while low RI indicates that no similar compounds with consistent data are present in the training set.
- Displays 5 most similar compounds from the training set for each considered aquatic species along with experimental toxicity values and similarities to the currently analyzed compound.
- The training sets used to build the models contain experimental data on aquatic toxicity to fathead minnows for about 900 compounds, to water fleas for about 600 compounds, and to ciliate protozoa for about 1100 compounds.
IMPORTANT NOTE:
If you installed Percepta as an upgrade over a previous version, the program will attempt to preserve any custom configuration of Self-training libraries used in Aquatic Toxicity module. This configuration will not include the new, significantly extended built-in libraries that were introduced in 2024 release. In this case, to take advantage of the new libraries, you may need to click "Configure" for the respective endpoint and manually select the following entries: LC50 (D. magna) v. 1.3 (read-only), IGC50 (T. pyriformis) v. 1.1 (read-only).
In case of a new installation, the new library should be selected automatically with no further action required.
Interface
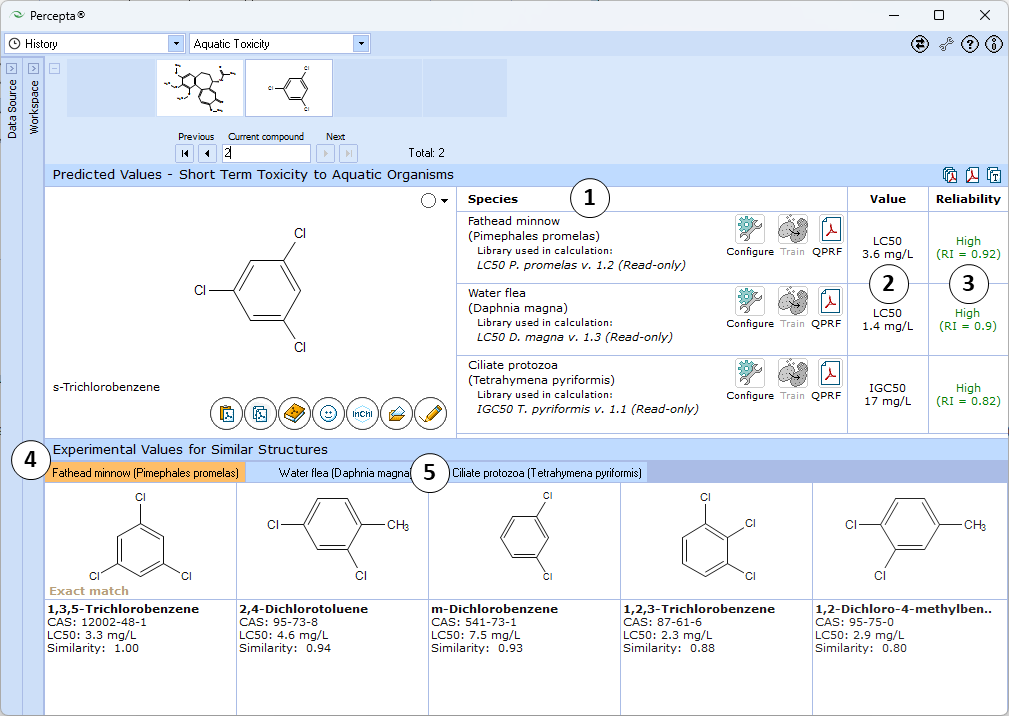
- Calculations are presented in the form of a table. Each row contains dedicated "Configure" and "Train" buttons to select the training library for the particular species and to add new data to that library, as well as "QPRF" button to generate the QPRF report in PDF format. Predictions are made for the three aquatic species most frequently used for testing - fathead minnows (Pimephales promelas), water fleas (Daphnia magna), and ciliate protozoa (Tetrahymena pyriformis).
- The predicted value is either LC50, or IGC50 of the analyzed compound to a given organism, expressed in mg/L.
- Predictions are supported by Reliability Index values ranging from 0 to 1 that serve as an intrinsic evaluation of prediction confidence:
- RI < 0.3 – Not Reliable,
- RI in range 0.3-0.5 – Bordeline Reliability,
- RI in range 0.5-0.75 – Moderate Reliability,
- RI >= 0.75 – High Reliability
- Up to five most similar compounds from the training set with names, CAS numbers and experimental LC50/IGC50 values.
- Click the tab to browse the similar structures for different species.
Technical information
Calculated quantitative parameters
The standard measure of aquatic toxicity is the concentration of the compound in water that is lethal to 50% of exposed organisms (LC50). In the case of protozoa, the endpoint is not lethality, but inhibition of cell growth by 50% (IGC50). To obtain a linear relationship with structural properties these data were converted to logarithmic form (pLC50/pIGC50) for modeling, but the final prediction result is returned as an original LC50/IGC50 value in mg/L.
Experimental data
Experimental data used for the development of predictive models were collected from EPA reference databases, as well as original publications. After thorough verification of the obtained values the final data sets contained about 900 compounds with quantitative LC50 values characterizing acute toxicity to fishes (Pimephales promelas), 600 compounds - to water fleas (Daphnia magna), and 1100 compounds - to ciliate protozoa (Tetrahymena pyriformis).
Model features & prediction accuracy
The predictive models were derived using GALAS (Global, Adjusted Locally According to Similarity) modeling methodology (please refer to [1] for more details).
Each GALAS model consists of two parts:
- Global (baseline) statistical model that reflects general trends in the variation of the property of interest.
- Similarity-based routine that performs local correction of baseline predictions taking into account the differences between baseline and experimental LC50 values for the most similar training set compounds.
GALAS methodology also provides the basis for estimating reliability of predictions by the means of calculated Reliability Index (RI) value that takes into account:
- Similarity of tested compound to the training set molecules.
- Consistence of experimental LC50 values and baseline model prediction for the most similar similar compounds from the training set.
Reliability Index ranges from 0 to 1 (0 corresponds to a completely unreliable, and 1 - a highly reliable prediction) and serves as an indication whether a submitted compound falls within the Model Applicability Domain. Compounds obtaining predictions RI < 0.3 are considered outside of the Applicability Domain of the model.
The resulting models are highly accurate: LC50 values for aquatic species are predicted with RMSE of 0.5-0.6 log units when only predictions of moderate and high reliability (RI >= 0.5) are considered .RI values in the high and moderate ranges are commonly obtained for 30-60% of the validation sets. Validation results also show that the accuracy of predictions is proportional to the Reliability Index, as shown in the table below for LC50 to fishes (P. promelas):
| Subset | Coverage of the entire internal validation set (N=175) |
R2 | RMSE | ||
|---|---|---|---|---|---|
| RI > 0.3 |
|
0.656 | 0.797 | ||
| RI > 0.5 |
|
0.795 | 0.501 | ||
| RI > 0.7 |
|
0.880 | 0.363 |
For more information regarding the modeling principles and validation results please refer to [2].