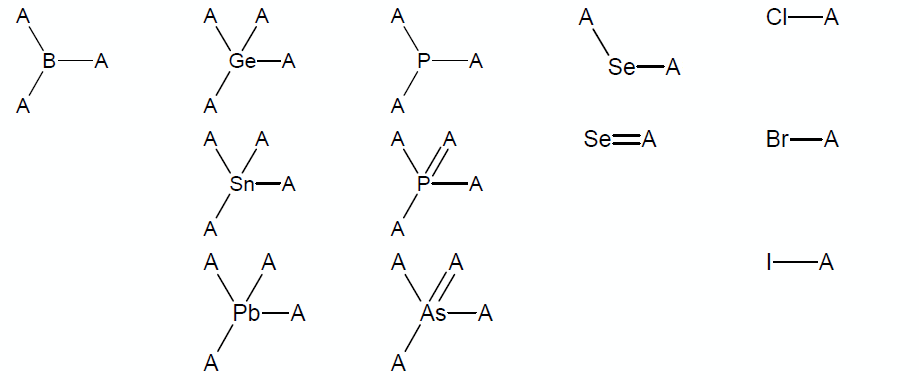LogP: Difference between revisions
Added new statistics |
|||
| (24 intermediate revisions by 2 users not shown) | |||
| Line 9: | Line 9: | ||
* Offers color-coded representation of lipophilic and hydrophilic parts of the compound structure. | * Offers color-coded representation of lipophilic and hydrophilic parts of the compound structure. | ||
* Train the model with experimental values to improve predictions for proprietary chemical space | * Train the model with experimental values to improve predictions for proprietary chemical space | ||
< | |||
<span style="color:red; font-weight: bold;">IMPORTANT NOTE:</span> | |||
If you installed Percepta as an upgrade over a previous version, the program will attempt to preserve any custom configuration of Self-training libraries used in ACD/LogP GALAS module. This configuration will not include the new, significantly extended built-in library that was introduced in 2024 release. In this case, to take advantage of the new library, you may need to click "Configure" and manually select the following entry: ''LogP v. 1.4 (read-only)''. | |||
In case of a new installation, the new library should be selected automatically with no further action required. | |||
== Interface == | == Interface == | ||
| Line 17: | Line 22: | ||
<br /> | <br /> | ||
[[Image: | [[Image:Acdlogp_classic.png|center]] | ||
<br /> | <br /> | ||
# LogP prediction obtained using ACD/LogP Classic calculation algorithm. | # LogP prediction obtained using ACD/LogP Classic calculation algorithm. | ||
| Line 29: | Line 34: | ||
<br /> | <br /> | ||
[[Image: | [[Image:Acdlogp_galas.png|center]] | ||
<br /> | <br /> | ||
# Lipophilic parts of the molecule are highlighted in green, hydrophilic groups in red, and the intensity of the color indicates the predicted degree of lipophilicity or hydrophilicity of an atom or a substructure. | # Lipophilic parts of the molecule are highlighted in green, hydrophilic groups in red, and the intensity of the color indicates the predicted degree of lipophilicity or hydrophilicity of an atom or a substructure. | ||
| Line 41: | Line 46: | ||
<br /> | <br /> | ||
[[Image: | [[Image:Logp_consensus.png|center]] | ||
<br /> | <br /> | ||
# ... | # The consensus LogP model predicts LogP as a weighted average of ACD/LogP Classic and ACD/LogP GALAS predictions. Each of the individual models is assigned with dynamic adaptive coefficients according to the indications of prediction quality. As a result, each model obtains larger weight in those regions of chemical space where it performs most reliably. The provided equation lists the weighting coefficients obtained for both models and the final Consensus LogP value. | ||
# | # Hover over the algorithm name in the displayed equation to view prediction details (calculated values, reliabilities and training options) from the underlying Classic and GALAS algorithms. | ||
# Shows 5 most similar compounds from '''LogP DB''' with experimental LogP values and literature references. The displayed similar structures are the same as in ACD/LogP GALAS module. | |||
<br /> | <br /> | ||
<div class="mw-collapsible | |||
<div id="Expand" style="display:inline;"></div> | |||
<div class="mw-collapsible"> | |||
==Technical information== | ==Technical information== | ||
<div class="mw-collapsible-content"> | <div class="mw-collapsible-content"> | ||
''' | ===Introduction to the 1-Octanol/Water Partitioning Coefficient=== | ||
''' | |||
'' | The octanol-water partition coefficient, log''P''<sub>o/w</sub>, is a measure of a compound’s hydrophobicity, which in many cases correlates well with various other properties of that compound, such as: | ||
* Extraction coefficients; | |||
* Retention on the reversed phase (RP) layers; | |||
* Transport and permeation through membranes; | |||
* Interaction with biological receptors and enzymes; | |||
* Toxicity; | |||
* Biological potency. | |||
Once you have obtained reliable log''P'' values for a series of compounds, you are able to estimate many of their properties that correlate with log''P''. | |||
===Database of Experimental LogP Values=== | |||
The main sources of experimental data, comprising the '''ACD/LogP DB''' were: | |||
* Reference books: | * Reference books: | ||
** <i>The Merck Index. An Encyclopedia of Chemicals, Drugs, and Biologicals</i>, O'Neil, M.J., Smith, A., Heckelman, P.E., Budavari, S., Eds. 13th Edition, Merck & Co., Inc., Whitehouse Station, NJ, 2001 | ** <i>The Merck Index. An Encyclopedia of Chemicals, Drugs, and Biologicals</i>, O'Neil, M.J., Smith, A., Heckelman, P.E., Budavari, S., Eds. 13th Edition, Merck & Co., Inc., Whitehouse Station, NJ, 2001 | ||
| Line 65: | Line 84: | ||
* Various articles from peer-reviewed scientific journals* | * Various articles from peer-reviewed scientific journals* | ||
* Other public data sources (online databases, handbooks, etc.) | * Other public data sources (online databases, handbooks, etc.) | ||
<span style="font-size:8pt"> | <span style="font-size:8pt"> | ||
<nowiki>*</nowiki> - Articles reporting LogP models by other authors were the predominant type among analyzed literature, meaning that each publication contained larger collections of experimental data (usually in the order of tens or hundreds compounds) compiled from corresponding original experimental articles. | <nowiki>*</nowiki> - Articles reporting LogP models by other authors were the predominant type among analyzed literature, meaning that each publication contained larger collections of experimental data (usually in the order of tens or hundreds compounds) compiled from corresponding original experimental articles. | ||
</span> | </span> | ||
< | |||
In '''ACD/Percepta''', the internal database is directly accessible and searchable under '''Databases\LogP''' data source, where each compound is provided with available experimental LogP values and references to the original literature. | |||
===Description of ACD/LogP GALAS Algorithm=== | |||
ACD/LogP GALAS module provides the estimate of the octanol-water partitioning coefficient for neutral species derived on the basis of GALAS (Global, Adjusted Locally According to Similarity) modeling methodology (please refer to [http://www.ncbi.nlm.nih.gov/pubmed/20373217] for more details). | |||
Each GALAS model consists of two parts: | |||
* Global (baseline) statistical model that reflects general trends in the variation of the property of interest. | |||
* Similarity-based routine that performs local correction of baseline predictions taking into account the differences between baseline and experimental LogP values for the most similar training set compounds. | |||
GALAS methodology also provides the basis for estimating reliability of predictions by the means of calculated Reliability Index (''RI'') value that takes into account: | |||
* Similarity of tested compound to the training set molecules. | |||
* Consistence of experimental LogP values and baseline model prediction for the most similar similar compounds from the training set. | |||
Reliability Index ranges from 0 to 1 (0 corresponds to a completely unreliable, and 1 - a highly reliable prediction) and serves as an indication whether a submitted compound falls within the Model Applicability Domain. Compounds obtaining predictions ''RI'' < 0.3 are considered outside of the Applicability Domain of the model. | |||
In addition, ACD/LogP GALAS algorithm provides a color-coded representation of the predicted property distribution indicating lipophilic and hydrophilic parts of the compound structure. | |||
====Internal Validation==== | ====Internal Validation==== | ||
Prior to model development, the compounds comprising the ACD/LogP DB were randomly split into a training set used for building the model, and a test set reserved for validation purposes: | |||
* '''Training set size:''' 11,387 | |||
* '''Internal validation set size:''' 4,890 | |||
Validation results are presented in the table below. | |||
{| cellpadding="2" cellspacing="0" style="border-top:2px solid black; border-bottom:2px solid black" | {| cellpadding="2" cellspacing="0" style="border-top:2px solid black; border-bottom:2px solid black" | ||
|+ <b>Table 1.</b> ACD/LogP | |+ <b>Table 1.</b> ACD/LogP GALAS model performance statistics for the various fractions of the internal validation set. | ||
|- | |- | ||
! style="border-bottom:1px solid black; background:#EAEAEA" width="150" | Subset | ! style="border-bottom:1px solid black; background:#EAEAEA" width="150" | Subset | ||
| Line 107: | Line 147: | ||
| align="center" | 0.96 || align="center" | 0.36 | | align="center" | 0.96 || align="center" | 0.36 | ||
|} | |} | ||
The statistics above apply to ACD/LogP GALAS algorithm with built-in "LogP v. 1.2" self-training library. Since then the library has undergone several updates. | |||
* "LogP v. 1.3" library introduced in 2018 has been expanded by >1700 drug-like compounds from novel congeneric series (representing a 12% increase in size) resulting in improved accuracy and reliability of prediction for novel entities: LogP was predicted within 0.5 log units for 85% of the novel compounds as demonstrated in the figures below: | |||
[[File:LogP_GALAS_v2017_vs_v2018.png|frame|none|Improvement in accuracy of logP prediction for the new set of 1724 compounds with the logP GALAS prediction model]] | |||
[[File:LogP_GALAS_v2017_to_v2018.png|frame|none|Improvements in logP prediction accuracy for the new set of 1724 compound from v2017.1 to v2018.1—less than 2% compounds have a prediction error greater than 1 logP unit in v 2018.1 compared to 30% in v2017.1]] | |||
* "LogP v. 1.4" library introduced in 2024 has been further expanded by ~4750 new compounds, resulting in a >25% increase over the previous version, and improving the prediction error for newly acquired compounds from ~1 to ~0.4 log units. | |||
[[File:LogP_GALAS_v2023_vs_v2024.png|frame|none|Improvement in accuracy of logP prediction for the new set of 4750 compounds with the logP GALAS prediction model]] | |||
===Description of ACD/LogP Classic Algorithm=== | |||
When a structure is entered for calculation, the program performs the following procedures: | |||
# Splits the structure into fragments. | |||
# Searches for identical fragments in the '''internal databases''': | |||
## The database of '''Fragmental Increments''' contains well-characterized increments for over 500 different functional groups. These differ from each other by their chemical structure (for example, amide, carboxy, ester, etc.), attachment to the hydrocarbon skeleton (aliphatic, vinylic, or aromatic), cyclization (cyclic or non-cyclic), and aromaticity (non-aromatic, aromatic, or fused aromatic). | |||
## The database of '''Carbon Atom Increments''' contains well-characterized increments for different types of carbons that are not involved in any functional group. They differ from each other by their state of hybridization (sp, sp2, or sp3), number of attached hydrogens, branching (primary, secondary, tertiary, or quaternary), cyclization (cyclic or non-cyclic), and aromaticity (non-aromatic, aromatic, or fused aromatic). | |||
## The database of the '''Intramolecular Interaction Increments''' contains well-characterized increments for over 2,000 different types of pair-wise group interactions. They differ from each other by the type of the interacting terminal groups (see the differences among functional groups above), and the length and type of the fragmental system between the interacting groups (aliphatic, aromatic, and vinylic). | |||
## Searches for identical fragments in the data sources specified for system training (for more information, refer to Appendix D). You can regulate the usage of each data field by its status: '''Included/Excluded''' in training, and statistical significance as '''High/Low'''. | |||
# If some fragments are not found in either of the above-mentioned databases, their increments (as well as increments of inter-fragmental interactions) are estimated using '''Secondary Algorithms'''. | |||
The algorithm estimates the probability of tautomeric and ionic equilibria, the calculation error, and displays the results. | |||
====Limitations==== | |||
ACD/LogP Classic algorithm does not calculate the logP values for the following chemical structures: | |||
* Charged structures other than the zwitterionic alpha-amino acids and their peptide derivatives, and other than the non-ionic derivatives of IV-valent Nitrogen (+) bonded to Oxygen (–) or bonded to Nitrogen (–) with double bond (note that such compounds can be calculated, if their logP values are included in the user database that is used for system training) | |||
* Structures containing atoms other than C, H, O, S, N, or F in possible chemical surroundings or structures containing atoms P, Cl, Br, I, Se, Si, Ge, Pb, Sn, As, or B that are not within the chemical surroundings shown below (note that A denotes any atom out of C, O, S, N, F, or any group listed below): | |||
[[File:Classic_Algorithm_Limitations.png]] | |||
* Structures that contain elements in their non-typical valence | |||
* Structures with coordinating bonds | |||
* Structures containing more than 255 atoms excluding hydrogen. | |||
'''Note:''' The groups from the table except sp<sup>3</sup>-hybridized selene cannot be a part of a cycle. | |||
The program does not take into account the specific features of different geometric isomers, stereoisomers, conformers, isotopes, and structures with non-covalent bonds. | |||
It predicts logP values so that in most cases the reliable experimental measurements lie within the calculated ±logP interval. However, it is still possible that some new chemical structures might possess new specific structural features, such as far-range non-covalent bonding, intra-molecular shielding, or inter-molecular association. In such cases, the discrepancy between a newly measured experimental value and the calculated ±logP interval might occur. | |||
'''Note:''' There certainly exist some structures that formally meet the aforementioned limitations, but cannot be calculated by the current algorithm. | |||
===Example of Structure-Fragment Approach=== | |||
[[File:LogP_Scheme.gif|center]] | |||
===When is Calculated Log''P'' More Accurate than Experimental?=== | |||
Bear in mind that log''P'' is a macroscopic measurement. The usefulness of the log''P'' parameter for many practical correlations is based on the assumption that '''log''P'' is a property of a single molecule'''. But in many cases, such a "thermodynamically pure" log''P'' value is very difficult, or even impossible to obtain experimentally. | |||
* The upper and lower limits for log''P'' values that can be measured by the traditional experimental procedures are ''ca.'' +8.0 and –3.0 respectively. It is very difficult to obtain reliable log''P'' values outside of this range. | |||
* In most cases, it is not possible to measure log''P'' values separately for all of the individual tautomeric forms. | |||
* There is no way to measure the exact log''P'' values for uncharged molecules of various amino acids, peptides, nucleosides, and any other compounds bearing both acidic (for example, -COOH or -PO<sub>3</sub>H<sub>2</sub>) and basic (for example, -NH<sub>2</sub>) groups. At any given pH, these molecules exist almost entirely in various ionic forms, and the concentration of non-ionized species of amino acids and similar forms is negligible. | |||
* Some compounds are '''unstable or nonexistent''' under the required conditions, for example, at extreme pH values which are necessary to suppress the acid-base equilibria. | |||
In all of these cases, the calculated log''P'' values are very likely to be of a greater reliability, compared to those measured experimentally. | |||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 13:35, 23 September 2024
Overview
This module calculates the value of the octanol-water partition coefficient – LogP. LogP predictions are exploited in many of our PhysChem and ADME prediction modules including LogD, Oral Bioavailability, Blood-Brain Barrier Permeation, and Passive Absorption, as well as in several Toxicity modules, such as hERG inhibition or Aquatic toxicity.
Features
- Includes two different predictive algorithms – ACD/LogP Classic and ACD/LogP GALAS. A Consensus logP based on these two models is also available.
- Provides a quantitative estimate of reliability of prediction by the means of 95% confidence intervals (ACD/LogP Classic), or Reliability Index (ACD/LogP GALAS).
- Offers color-coded representation of lipophilic and hydrophilic parts of the compound structure.
- Train the model with experimental values to improve predictions for proprietary chemical space
IMPORTANT NOTE:
If you installed Percepta as an upgrade over a previous version, the program will attempt to preserve any custom configuration of Self-training libraries used in ACD/LogP GALAS module. This configuration will not include the new, significantly extended built-in library that was introduced in 2024 release. In this case, to take advantage of the new library, you may need to click "Configure" and manually select the following entry: LogP v. 1.4 (read-only).
In case of a new installation, the new library should be selected automatically with no further action required.
Interface
ACD/LogP Classic
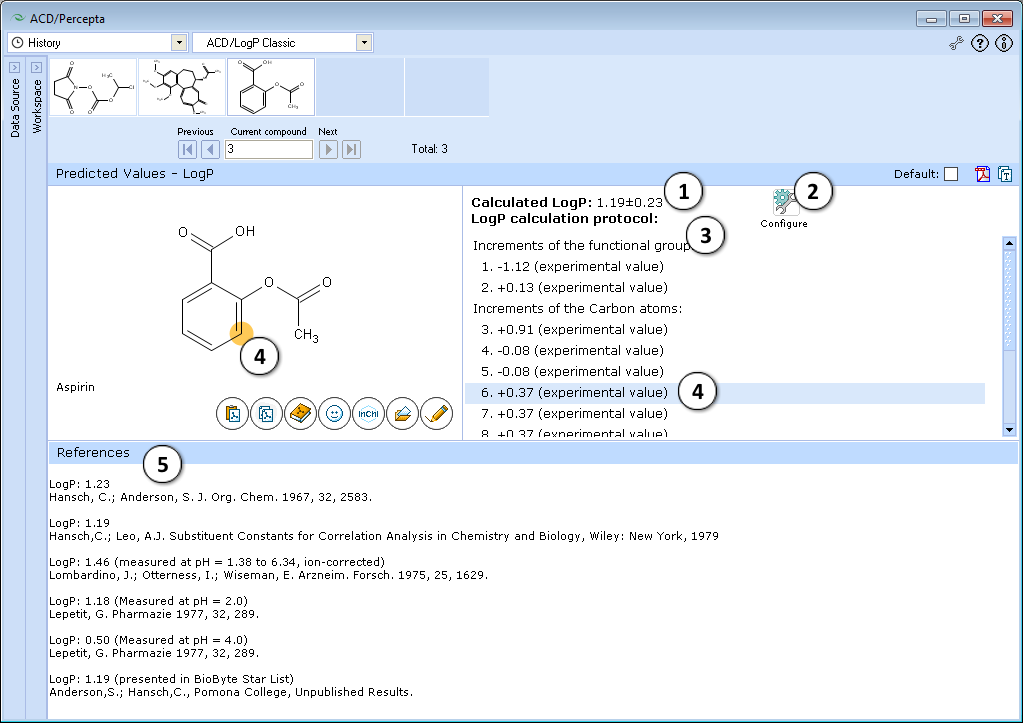
- LogP prediction obtained using ACD/LogP Classic calculation algorithm.
- Press "Configure" button to switch model training on or off, and to select the database file to use for training.
- LogP calculation protocol. Lists the increments of all functional groups and carbon atoms, as well as the contirbutions of interaction through aliphatic, aromatic and vinylic systems.
- The protocol is interactive. Click on any entry to highlight the respective atom, group, or interaction onto the molecule.
- If the compound is found in LogP DB, all available experimental data for that compound are displayed along with literature references.
ACD/LogP GALAS
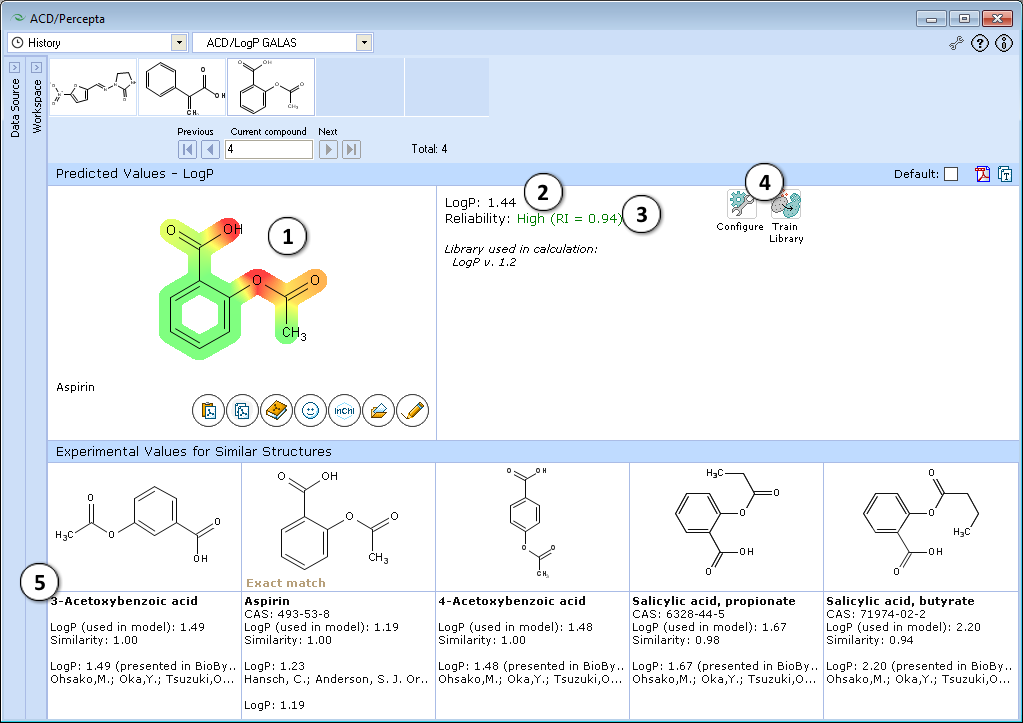
- Lipophilic parts of the molecule are highlighted in green, hydrophilic groups in red, and the intensity of the color indicates the predicted degree of lipophilicity or hydrophilicity of an atom or a substructure.
- LogP prediction obtained using ACD/LogP GALAS calculation algorithm.
- Reliability index (RI):
RI < 0.3 – Not Reliable,
RI in range 0.3-0.5 – Borderline Reliability,
RI in range 0.5-0.75 – Moderate Reliability,
RI >= 0.75 – High Reliability - "Configure" and "Train" buttons provide the means to select the training library for use in calculations and to add new data to that library. The name of the currently selected library is indicated with italic font.
- Displays 5 most similar compounds from LogP DB with experimental LogP values and literature references
Consensus LogP
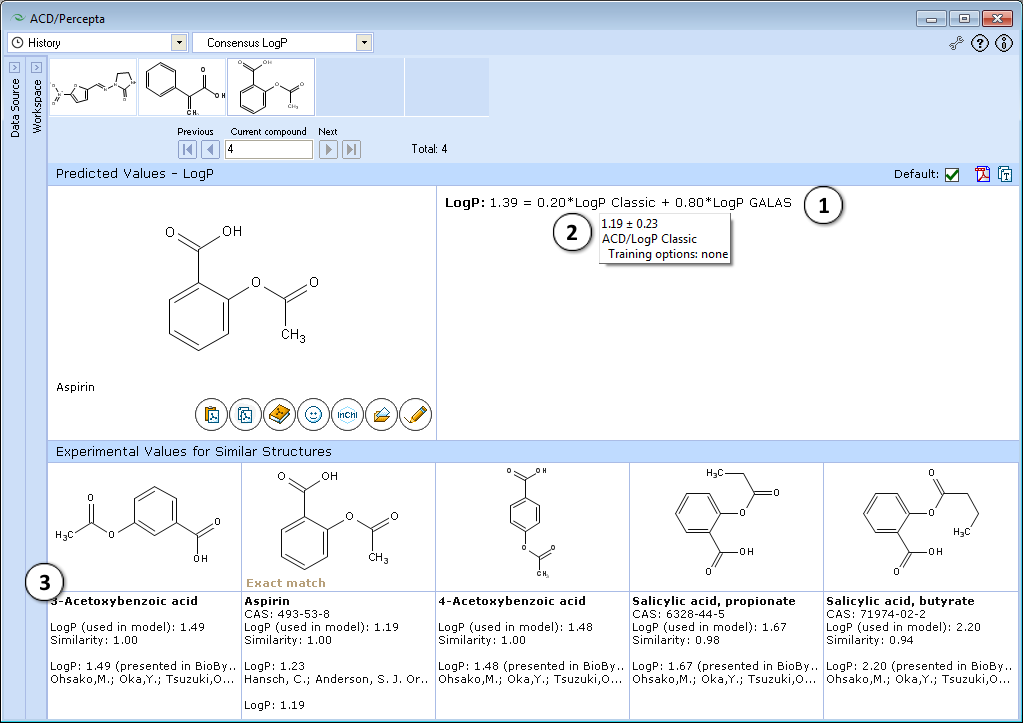
- The consensus LogP model predicts LogP as a weighted average of ACD/LogP Classic and ACD/LogP GALAS predictions. Each of the individual models is assigned with dynamic adaptive coefficients according to the indications of prediction quality. As a result, each model obtains larger weight in those regions of chemical space where it performs most reliably. The provided equation lists the weighting coefficients obtained for both models and the final Consensus LogP value.
- Hover over the algorithm name in the displayed equation to view prediction details (calculated values, reliabilities and training options) from the underlying Classic and GALAS algorithms.
- Shows 5 most similar compounds from LogP DB with experimental LogP values and literature references. The displayed similar structures are the same as in ACD/LogP GALAS module.
Technical information
Introduction to the 1-Octanol/Water Partitioning Coefficient
The octanol-water partition coefficient, logPo/w, is a measure of a compound’s hydrophobicity, which in many cases correlates well with various other properties of that compound, such as:
- Extraction coefficients;
- Retention on the reversed phase (RP) layers;
- Transport and permeation through membranes;
- Interaction with biological receptors and enzymes;
- Toxicity;
- Biological potency.
Once you have obtained reliable logP values for a series of compounds, you are able to estimate many of their properties that correlate with logP.
Database of Experimental LogP Values
The main sources of experimental data, comprising the ACD/LogP DB were:
- Reference books:
- The Merck Index. An Encyclopedia of Chemicals, Drugs, and Biologicals, O'Neil, M.J., Smith, A., Heckelman, P.E., Budavari, S., Eds. 13th Edition, Merck & Co., Inc., Whitehouse Station, NJ, 2001
- Therapeutic Drugs, Dolery, C., Ed. 2nd Edition, Churchill Livingstone, New York, NY, 1999
- Clarke's Isolation and Identification of Drugs, Moffat, A.C., Jackson, J.V., Moss, M.S., Widdop, B., Eds. 2nd Edition, The Pharmaceutical Press, London, 1986
- Various articles from peer-reviewed scientific journals*
- Other public data sources (online databases, handbooks, etc.)
* - Articles reporting LogP models by other authors were the predominant type among analyzed literature, meaning that each publication contained larger collections of experimental data (usually in the order of tens or hundreds compounds) compiled from corresponding original experimental articles.
In ACD/Percepta, the internal database is directly accessible and searchable under Databases\LogP data source, where each compound is provided with available experimental LogP values and references to the original literature.
Description of ACD/LogP GALAS Algorithm
ACD/LogP GALAS module provides the estimate of the octanol-water partitioning coefficient for neutral species derived on the basis of GALAS (Global, Adjusted Locally According to Similarity) modeling methodology (please refer to [1] for more details).
Each GALAS model consists of two parts:
- Global (baseline) statistical model that reflects general trends in the variation of the property of interest.
- Similarity-based routine that performs local correction of baseline predictions taking into account the differences between baseline and experimental LogP values for the most similar training set compounds.
GALAS methodology also provides the basis for estimating reliability of predictions by the means of calculated Reliability Index (RI) value that takes into account:
- Similarity of tested compound to the training set molecules.
- Consistence of experimental LogP values and baseline model prediction for the most similar similar compounds from the training set.
Reliability Index ranges from 0 to 1 (0 corresponds to a completely unreliable, and 1 - a highly reliable prediction) and serves as an indication whether a submitted compound falls within the Model Applicability Domain. Compounds obtaining predictions RI < 0.3 are considered outside of the Applicability Domain of the model.
In addition, ACD/LogP GALAS algorithm provides a color-coded representation of the predicted property distribution indicating lipophilic and hydrophilic parts of the compound structure.
Internal Validation
Prior to model development, the compounds comprising the ACD/LogP DB were randomly split into a training set used for building the model, and a test set reserved for validation purposes:
- Training set size: 11,387
- Internal validation set size: 4,890
Validation results are presented in the table below.
| Subset | Coverage of the entire internal validation set (N=4,890) |
R2 | RMSE | ||
|---|---|---|---|---|---|
| RI > 0.3 N = 4,872 |
|
0.94 | 0.46 | ||
| RI > 0.5 N = 4,772 |
|
0.95 | 0.44 | ||
| RI > 0.75 N = 3,345 |
|
0.96 | 0.36 |
The statistics above apply to ACD/LogP GALAS algorithm with built-in "LogP v. 1.2" self-training library. Since then the library has undergone several updates.
- "LogP v. 1.3" library introduced in 2018 has been expanded by >1700 drug-like compounds from novel congeneric series (representing a 12% increase in size) resulting in improved accuracy and reliability of prediction for novel entities: LogP was predicted within 0.5 log units for 85% of the novel compounds as demonstrated in the figures below:
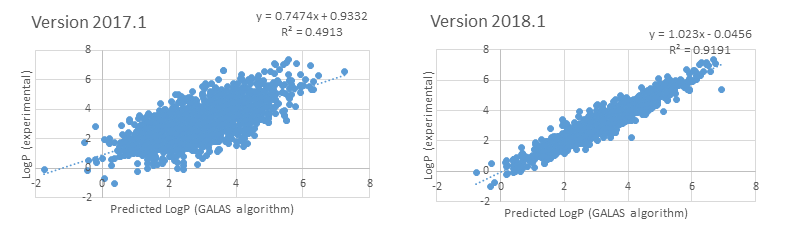
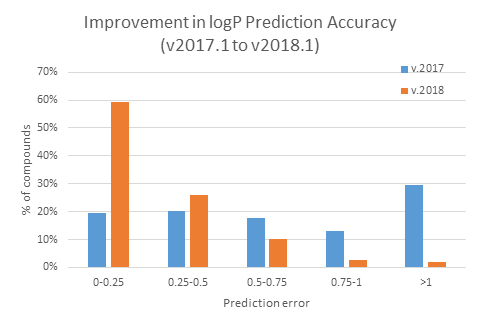
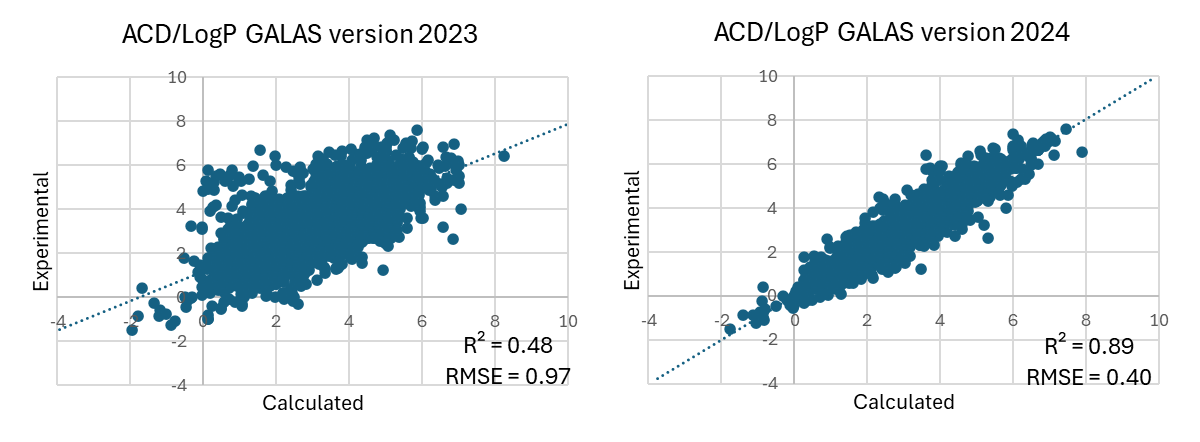
- "LogP v. 1.4" library introduced in 2024 has been further expanded by ~4750 new compounds, resulting in a >25% increase over the previous version, and improving the prediction error for newly acquired compounds from ~1 to ~0.4 log units.
Description of ACD/LogP Classic Algorithm
When a structure is entered for calculation, the program performs the following procedures:
- Splits the structure into fragments.
- Searches for identical fragments in the internal databases:
- The database of Fragmental Increments contains well-characterized increments for over 500 different functional groups. These differ from each other by their chemical structure (for example, amide, carboxy, ester, etc.), attachment to the hydrocarbon skeleton (aliphatic, vinylic, or aromatic), cyclization (cyclic or non-cyclic), and aromaticity (non-aromatic, aromatic, or fused aromatic).
- The database of Carbon Atom Increments contains well-characterized increments for different types of carbons that are not involved in any functional group. They differ from each other by their state of hybridization (sp, sp2, or sp3), number of attached hydrogens, branching (primary, secondary, tertiary, or quaternary), cyclization (cyclic or non-cyclic), and aromaticity (non-aromatic, aromatic, or fused aromatic).
- The database of the Intramolecular Interaction Increments contains well-characterized increments for over 2,000 different types of pair-wise group interactions. They differ from each other by the type of the interacting terminal groups (see the differences among functional groups above), and the length and type of the fragmental system between the interacting groups (aliphatic, aromatic, and vinylic).
- Searches for identical fragments in the data sources specified for system training (for more information, refer to Appendix D). You can regulate the usage of each data field by its status: Included/Excluded in training, and statistical significance as High/Low.
- If some fragments are not found in either of the above-mentioned databases, their increments (as well as increments of inter-fragmental interactions) are estimated using Secondary Algorithms.
The algorithm estimates the probability of tautomeric and ionic equilibria, the calculation error, and displays the results.
Limitations
ACD/LogP Classic algorithm does not calculate the logP values for the following chemical structures:
- Charged structures other than the zwitterionic alpha-amino acids and their peptide derivatives, and other than the non-ionic derivatives of IV-valent Nitrogen (+) bonded to Oxygen (–) or bonded to Nitrogen (–) with double bond (note that such compounds can be calculated, if their logP values are included in the user database that is used for system training)
- Structures containing atoms other than C, H, O, S, N, or F in possible chemical surroundings or structures containing atoms P, Cl, Br, I, Se, Si, Ge, Pb, Sn, As, or B that are not within the chemical surroundings shown below (note that A denotes any atom out of C, O, S, N, F, or any group listed below):
- Structures that contain elements in their non-typical valence
- Structures with coordinating bonds
- Structures containing more than 255 atoms excluding hydrogen.
Note: The groups from the table except sp3-hybridized selene cannot be a part of a cycle.
The program does not take into account the specific features of different geometric isomers, stereoisomers, conformers, isotopes, and structures with non-covalent bonds.
It predicts logP values so that in most cases the reliable experimental measurements lie within the calculated ±logP interval. However, it is still possible that some new chemical structures might possess new specific structural features, such as far-range non-covalent bonding, intra-molecular shielding, or inter-molecular association. In such cases, the discrepancy between a newly measured experimental value and the calculated ±logP interval might occur.
Note: There certainly exist some structures that formally meet the aforementioned limitations, but cannot be calculated by the current algorithm.
Example of Structure-Fragment Approach
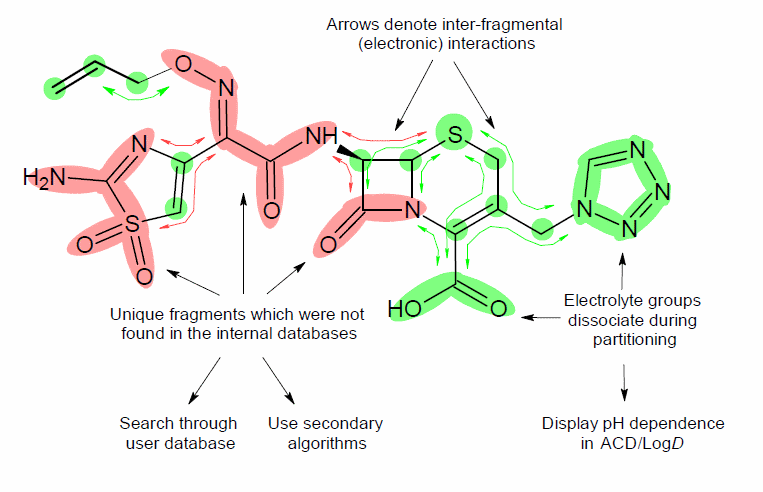
When is Calculated LogP More Accurate than Experimental?
Bear in mind that logP is a macroscopic measurement. The usefulness of the logP parameter for many practical correlations is based on the assumption that logP is a property of a single molecule. But in many cases, such a "thermodynamically pure" logP value is very difficult, or even impossible to obtain experimentally.
- The upper and lower limits for logP values that can be measured by the traditional experimental procedures are ca. +8.0 and –3.0 respectively. It is very difficult to obtain reliable logP values outside of this range.
- In most cases, it is not possible to measure logP values separately for all of the individual tautomeric forms.
- There is no way to measure the exact logP values for uncharged molecules of various amino acids, peptides, nucleosides, and any other compounds bearing both acidic (for example, -COOH or -PO3H2) and basic (for example, -NH2) groups. At any given pH, these molecules exist almost entirely in various ionic forms, and the concentration of non-ionized species of amino acids and similar forms is negligible.
- Some compounds are unstable or nonexistent under the required conditions, for example, at extreme pH values which are necessary to suppress the acid-base equilibria.
In all of these cases, the calculated logP values are very likely to be of a greater reliability, compared to those measured experimentally.