hERG Inhibition: Difference between revisions
No edit summary |
|||
| (6 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<br /> | <br /> | ||
Cardiotoxicity of drug-like compounds associated with human ether-a-go-go (hERG) channel inhibition is becoming more and more common cause of drug candidates’ attrition. The hERG potassium channel is required for normal cardiac depolarization and its blockage can lead to cardiac QT interval prolongation and life threatening arrhythmias. | Cardiotoxicity of drug-like compounds associated with human ether-a-go-go (hERG) channel inhibition is becoming more and more common cause of drug candidates’ attrition. The hERG potassium channel is required for normal cardiac depolarization and its blockage can lead to cardiac QT interval prolongation and life-threatening arrhythmias. | ||
Using hERG inhibition module, you have the capability to quickly identify hERG inhibitors. Training of models using usually very large ‘in-house’ experimental (screening) data of hERG inhibition would expand the Applicability Domain of the model and would produce reliable predictions for compounds synthesized in your company. Moreover, training allows customization of our model to ensure that it correctly handles the data originating from the | Using hERG inhibition module, you have the capability to quickly identify hERG inhibitors. Training of models using usually very large ‘in-house’ experimental (screening) data of hERG inhibition would expand the Applicability Domain of the model and would produce reliable predictions for compounds synthesized in your company. Moreover, training allows customization of our model to ensure that it correctly handles the data originating from the screening protocol used in your company that may significantly differ from standard protocols described in the literature. | ||
<br /> | <br /> | ||
===Features=== | ===Features=== | ||
* Predicts the probability for a compound to inhibit hERG channel at clinically relevant concentrations (K<sub>i</sub> < 10 μM). | * Predicts the probability for a compound to inhibit hERG channel at clinically relevant concentrations (K<sub>i</sub> < 10 μM). | ||
* | * Predictions are based on a data set of almost 9400 compounds with experimental results collected from published hERG inhibition studies utilizing either patch-clamp or competitive binding methods. | ||
* Calculates Reliability Index (RI values) of predictions that indicates whether tested compounds belong to Applicability Domain of predictive model. | * Calculates Reliability Index (RI values) of predictions that indicates whether tested compounds belong to Applicability Domain of predictive model. | ||
* Performs a similarity search and displays top 5 most similar structures from the training set of the model along with their names, experimental results, and literature references. | * Performs a similarity search and displays top 5 most similar structures from the training set of the model along with their names, experimental results, and literature references. | ||
* | * Supports training the model using ‘in-house’ data, including those generated by ‘in-house’ screening protocol. | ||
* [[hERG_Inhibition#hERG_IC50|hERG_IC50]] module enables exploration of the influence of key physicochemical properties of drugs on their hERG liability and provides a quantitative estimate of inhibitory potency in the form of predicted IC50 value. | |||
<span style="color:red; font-weight: bold;">IMPORTANT NOTE:</span> | <span style="color:red; font-weight: bold;">IMPORTANT NOTE:</span> | ||
If you installed | If you installed Percepta as an in-place upgrade with Expert user privileges, hERG Inhibition module will continue using the same set of Self-training libraries that was configured in your previous installation. If you are upgrading over version 2021 or earlier, this will not include the new, significantly extended built-in library that comes with the software since v. 2022. To take advantage of this library, you will need to click "Configure" and manually select the following entry: ''hERG-I (Ki less than 10 uM) v. 1.4 (read-only)''. | ||
In case of a clean installation, this library is selected automatically, and no further action is required. | In case of a clean installation, or an upgrade with a Limited user key, this library is selected automatically, and no further action is required. | ||
<br /> | <br /> | ||
| Line 24: | Line 25: | ||
<br /> | <br /> | ||
===hERG Inhibitors=== | |||
[[Image:Herg_inhibition.png|center]] | [[Image:Herg_inhibition.png|center]] | ||
<br /> | <br /> | ||
# Estimated probability of | # Estimated probability of the compound being a hERG channel inhibitor. | ||
# Indication of the prediction reliability along with the Reliability Index value: | # Indication of the prediction reliability along with the Reliability Index value: | ||
#* RI < 0.3 – Not Reliable, | #* RI < 0.3 – Not Reliable, | ||
| Line 33: | Line 35: | ||
#* RI in range 0.5-0.75 – Moderate Reliability, | #* RI in range 0.5-0.75 – Moderate Reliability, | ||
#* RI >= 0.75 – High Reliability | #* RI >= 0.75 – High Reliability | ||
# | # "Configure" and "Train" buttons provide the means to select the training library for use in calculations and to add new data to that library. The names of the currently selected libraries are indicated with italic font | ||
# 5 most similar structures from the self-training library with compound names, experimental classification (Inhibitor or Non-inhibitor), and references | |||
<br /> | <br /> | ||
<div class="mw-collapsible | ===hERG IC50=== | ||
[[Image:Herg_IC50.png|center]] | |||
<br /> | |||
# Main physicochemical descriptors (LogP, Molecular Weight, Acid and Base pKa) that are used as inputs for calculation. By default, automatically calculated values are used. Enter experimental values to improve prediction accuracy or type custom values to model the influence of the respective properties on hERG inhibitory potential of the compound of interest. | |||
# Click the "Undo" button to revert the manually entered property value to the automatically calculated value (LogP and pKa (Acid) in this picture) for a compound and to recalculate hERG inhibition potency | |||
# Click to recalculate hERG inhibition potency using the currently specified parameter values | |||
# Estimated hERG half-inhibitory constant (IC<sub>50</sub>) of the compound | |||
# 5 most similar structures from the training set with compound names, experimental results (exact IC50 values or intervals), type of assay used for meaurements, and references | |||
# A "heatmap" plot illustrating the partial dependences of hERG inhibition potential on lipophilicity and ionization parameters. | |||
[[Image:Herg_heatmap.png|left]] | |||
:a. The cyan-colored dot indicates the position of the current compound. | |||
:b. Click to select the variables to plot: | |||
* LogP vs pKa (Acid) | |||
* LogP vs pKa (Base) | |||
* pKa (Acid) vs pKa (Base) | |||
<br style="clear:both"> | |||
<div class="mw-collapsible"> | |||
==Technical information== | ==Technical information== | ||
| Line 42: | Line 68: | ||
<div class="mw-collapsible-content"> | <div class="mw-collapsible-content"> | ||
===Experimental data=== | ===Experimental data & predictive models=== | ||
The current iteration of the built-in library included in hERG inhibition module contains 9383 compounds with experimental values determined using patch-clamp (conventional and automatic) and competitive radioligand displacement assays (reference ligands: dofetilide, astemizole, MK-499). The data were collected from ChEMBL database, as well as original literature publications. Detailed information about the employed data collection and processing procedures can be found in ''J Comput Aided Mol Des.'' '''2016''';30(12):1175-1188. [https://doi.org/10.1007/s10822-016-9986-0] | |||
* | |||
hERG Inhibition module group contains two different models: | |||
* hERG IC50 module presents a PhysChem-based quantitative model - a Gradient Boosting AFT (Accelerated Failure Time) model that has been trained using both fully quantitative and censored (interval) data. This model predicts predicts IC<sub>50</sub> values from a minimal set of physicochemical descriptors including octanol/water LogP, acid and base pKa, molecular size, topology and flexibility parameters. Technical information about the development procedures and performance of this model is available in ''J Comput Aided Mol Des.'' '''2022''';36(12):837-849. [https://doi.org/10.1007/s10822-022-00483-0] | |||
* hERG Inhibitors module provides a probabilistic GALAS model, which is based on a two-part trainable approach, involving a 'baseline' statistical fragmental model, and a similarity-based correction routine that forms the basis of trainability. Originally, this model was built using a smaller data set of 663 molecules with high quality quantitative data. The information provided below applies to that initial model. In the current version of the software, the original 'baseline' model has been trained using the full database of 9383 molecules to ensure the best possible coverage of pharmaceutically relevant chemical space. | |||
=== | ===Description of the original GALAS model=== | ||
====Data Conversion==== | |||
The following criteria were applied for conversion of continuous data representing strength of compounds' interaction with hERG channel to binary representation: | The following criteria were applied for conversion of continuous data representing strength of compounds' interaction with hERG channel to binary representation: | ||
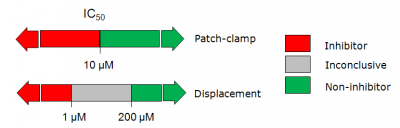
* In '''patch-clamp''' studies compounds that exhibited IC50 < | * In '''patch-clamp''' studies compounds that exhibited IC50 < 10 μM were considered hERG inhibitors, while those with IC50 > 10 μM – hERG non-inhibitors. | ||
* For the data coming from '''radioligand displacement assay''' the corresponding thresholds were as follows: ''K<sub>i</sub>'' < 0.5 μM - inhibitors, ''K<sub>i</sub>'' > 100 μM - non-inhibitors, while compounds in the intermediate range (0.5 μM < ''K<sub>i</sub>'' < 100 μM) were labeled inconclusive. | * For the data coming from '''radioligand displacement assay''' the corresponding thresholds were as follows: ''K<sub>i</sub>'' < 0.5 μM - inhibitors, ''K<sub>i</sub>'' > 100 μM - non-inhibitors, while compounds in the intermediate range (0.5 μM < ''K<sub>i</sub>'' < 100 μM) were labeled inconclusive. | ||
More strict criteria were applied to radioligand displacement data compared to patch-clamp studies since the former method does not provide a direct measure of hERG channel inhibition, but rather represents hERG binding affinity. To ensure high quality of the data set only sufficiently strong or weak binders were considered inhibitors or non-inhibitors respectively, while no definitive categories were | More strict criteria were applied to radioligand displacement data compared to patch-clamp studies since the former method does not provide a direct measure of hERG channel inhibition, but rather represents hERG binding affinity. To ensure high quality of the data set only sufficiently strong or weak binders were considered inhibitors or non-inhibitors respectively, while no definitive categories were assigned to compounds with moderate binding affinities. | ||
[[File:Herg_scale.png|400px]] | [[File:Herg_scale.png|400px]] | ||
===Model features & prediction accuracy=== | ====Model features & prediction accuracy==== | ||
Full methodological details of GALAS (Global, Adjusted Locally According to Similarity) modeling approach are available in ''SAR QSAR Environ Res.'' '''2010''';21(1):127-48. [https://doi.org/10.1080/10629360903568671] | |||
Each GALAS model consists of two parts: | Each GALAS model consists of two parts: | ||
* Global baseline statistical model employing binomial PLS with multiple bootstrapping using a predefined set of fragmental descriptors, that reflects general trends in | * Global baseline statistical model employing binomial PLS with multiple bootstrapping using a predefined set of fragmental descriptors, that reflects general trends in hERG inhibitory potential. | ||
* Similarity-based routine that performs local correction of baseline predictions | * Similarity-based routine that performs local correction of baseline predictions considering the differences between baseline and experimental values for the most similar training set compounds. | ||
<br> | <br> | ||
GALAS methodology also provides the basis for estimating reliability of predictions by the means of calculated Reliability Index (''RI'') value ranging from 0 to 1 that | GALAS methodology also provides the basis for estimating reliability of predictions by the means of calculated Reliability Index (''RI'') value ranging from 0 to 1 that considers the following two criteria: | ||
* Similarity of tested compound to the training set molecules (prediction is unreliable if no similar compounds have been found). | * Similarity of tested compound to the training set molecules (prediction is unreliable if no similar compounds have been found). | ||
* Consistence of experimental values and baseline model prediction for the most | * Consistence of experimental values and baseline model prediction for the most similar compounds from the training set (discrepant data for similar molecules, i.e. alternating hERG blockers and hERG non-blockers lead to lower ''RI'' values). | ||
The used method also provides the basis of model Trainability. 'Trainable model' methodology addresses the issue of the chemical space of ‘in-house’ libraries being considerably wider than that of publicly available data which results in limited applicability of most third-party QSARs for analysis of ‘in-house’ data. The ‘Training engine‘ makes appropriate corrections for systematic deviations produced by the baseline QSAR model based on analysis of similar compounds from the experimental data library. Expansion of this Self-training Library with user-defined experimental data for new compounds leads to instant improvement of prediction accuracy for the respective compound classes. Moreover, addition of 'in-house' data allows adapting the existing model to the particular experimental protocol used in your company and avoiding potential issues related to discrepancies between different experimental methods used for determination of drug interactions with hERG (see [[hERG_Inhibition#Model_Trainability_Demonstration|Model Trainability Demonstration]]) section. | The used method also provides the basis of model Trainability. 'Trainable model' methodology addresses the issue of the chemical space of ‘in-house’ libraries being considerably wider than that of publicly available data which results in limited applicability of most third-party QSARs for analysis of ‘in-house’ data. The ‘Training engine‘ makes appropriate corrections for systematic deviations produced by the baseline QSAR model based on analysis of similar compounds from the experimental data library. Expansion of this Self-training Library with user-defined experimental data for new compounds leads to instant improvement of prediction accuracy for the respective compound classes. Moreover, addition of 'in-house' data allows adapting the existing model to the particular experimental protocol used in your company and avoiding potential issues related to discrepancies between different experimental methods used for determination of drug interactions with hERG (see [[hERG_Inhibition#Model_Trainability_Demonstration|Model Trainability Demonstration]]) section. | ||
| Line 97: | Line 126: | ||
* Only compounds within Applicability Domain (RI > 0.3) were considered in testing. | * Only compounds within Applicability Domain (RI > 0.3) were considered in testing. | ||
===Model Trainability Demonstration=== | ====Model Trainability Demonstration==== | ||
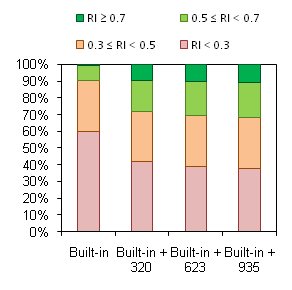
[[Image:HERG_PubChem.gif|right|Distribution of Test Set compound by RI values of predictions after addition of different portions of PubChem data set to the Self-training Library]] | [[Image:HERG_PubChem.gif|right|Distribution of Test Set compound by RI values of predictions after addition of different portions of PubChem data set to the Self-training Library]] | ||
Trainability of the described predictive model of hERG inhibition was tested using an external data set derived from HTS fluorescence assay that has recently become available in the PubChem database. Validation procedure was performed as follows: | Trainability of the described predictive model of hERG inhibition was tested using an external data set derived from HTS fluorescence assay that has recently become available in the PubChem database. Validation procedure was performed as follows: | ||
* HTS fluorescence assay data for 1609 compounds were extracted from Pubchem database. Quantitative values provided in the PubChem database (PubChem scores - fluorescence increase over negative control compared to reference compound terfenadine) were converted to binary representation: compounds with Pubchem score > 40% were considered hERG inhibitors; those with Pubchem score from -20 to 20% - non-inhibitors. | * HTS fluorescence assay data for 1609 compounds were extracted from Pubchem database. Quantitative values provided in the PubChem database (PubChem scores - fluorescence increase over negative control compared to reference compound terfenadine) were converted to binary representation: compounds with Pubchem score > 40% were considered hERG inhibitors; those with Pubchem score from -20 to 20% - non-inhibitors. | ||
* Part of this external data library was reserved as a test set. The remaining data were added to the | * Part of this external data library was reserved as a test set. The remaining data were added to the Self-training Library in three steps. | ||
Library in three steps. | |||
* The resulting models containing different portions of HTS data were validated against the reserved test set. | * The resulting models containing different portions of HTS data were validated against the reserved test set. | ||
When calculations for the test set are made using Built-in Self-training Library, predicted values for many compounds | When calculations for the test set are made using Built-in Self-training Library, predicted values for many compounds are marked ‘Not reliable’ (i.e. fall outside of the Model Applicability Domain, red bars in the figure). However, as discussed above, prediction accuracy is still impressive if calculations of at least borderline reliability (RI ≥ 0.3) are considered. The key point is the appearance of a considerable number of moderate (RI ≥ 0.5) and high-quality predictions (RI ≥ 0.7) when | ||
even a small part of external data set is added to the Self-training Library (green bars in the figure). The percentage of reliable predictions goes even higher with further expansion of the Library, while the same or better overall accuracy of calculations is maintained: | even a small part of external data set is added to the Self-training Library (green bars in the figure). The percentage of reliable predictions goes even higher with further expansion of the Library, while the same or better overall accuracy of calculations is maintained: | ||
| Line 134: | Line 162: | ||
|} | |} | ||
These results demonstrate the ability of our ‘Trainable model’ methodology to adapt the existing model to the | These results demonstrate the ability of our ‘Trainable model’ methodology to adapt the existing model to the chemical space represented by an external compound set. It is also obvious that our Training engine successfully corrects for the differences in experimental estimation when data from different assays are combined and therefore, is particularly suitable for analysis of ‘in-house’ data. | ||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 09:45, 26 July 2023
Overview
Cardiotoxicity of drug-like compounds associated with human ether-a-go-go (hERG) channel inhibition is becoming more and more common cause of drug candidates’ attrition. The hERG potassium channel is required for normal cardiac depolarization and its blockage can lead to cardiac QT interval prolongation and life-threatening arrhythmias.
Using hERG inhibition module, you have the capability to quickly identify hERG inhibitors. Training of models using usually very large ‘in-house’ experimental (screening) data of hERG inhibition would expand the Applicability Domain of the model and would produce reliable predictions for compounds synthesized in your company. Moreover, training allows customization of our model to ensure that it correctly handles the data originating from the screening protocol used in your company that may significantly differ from standard protocols described in the literature.
Features
- Predicts the probability for a compound to inhibit hERG channel at clinically relevant concentrations (Ki < 10 μM).
- Predictions are based on a data set of almost 9400 compounds with experimental results collected from published hERG inhibition studies utilizing either patch-clamp or competitive binding methods.
- Calculates Reliability Index (RI values) of predictions that indicates whether tested compounds belong to Applicability Domain of predictive model.
- Performs a similarity search and displays top 5 most similar structures from the training set of the model along with their names, experimental results, and literature references.
- Supports training the model using ‘in-house’ data, including those generated by ‘in-house’ screening protocol.
- hERG_IC50 module enables exploration of the influence of key physicochemical properties of drugs on their hERG liability and provides a quantitative estimate of inhibitory potency in the form of predicted IC50 value.
IMPORTANT NOTE:
If you installed Percepta as an in-place upgrade with Expert user privileges, hERG Inhibition module will continue using the same set of Self-training libraries that was configured in your previous installation. If you are upgrading over version 2021 or earlier, this will not include the new, significantly extended built-in library that comes with the software since v. 2022. To take advantage of this library, you will need to click "Configure" and manually select the following entry: hERG-I (Ki less than 10 uM) v. 1.4 (read-only).
In case of a clean installation, or an upgrade with a Limited user key, this library is selected automatically, and no further action is required.
Interface
hERG Inhibitors
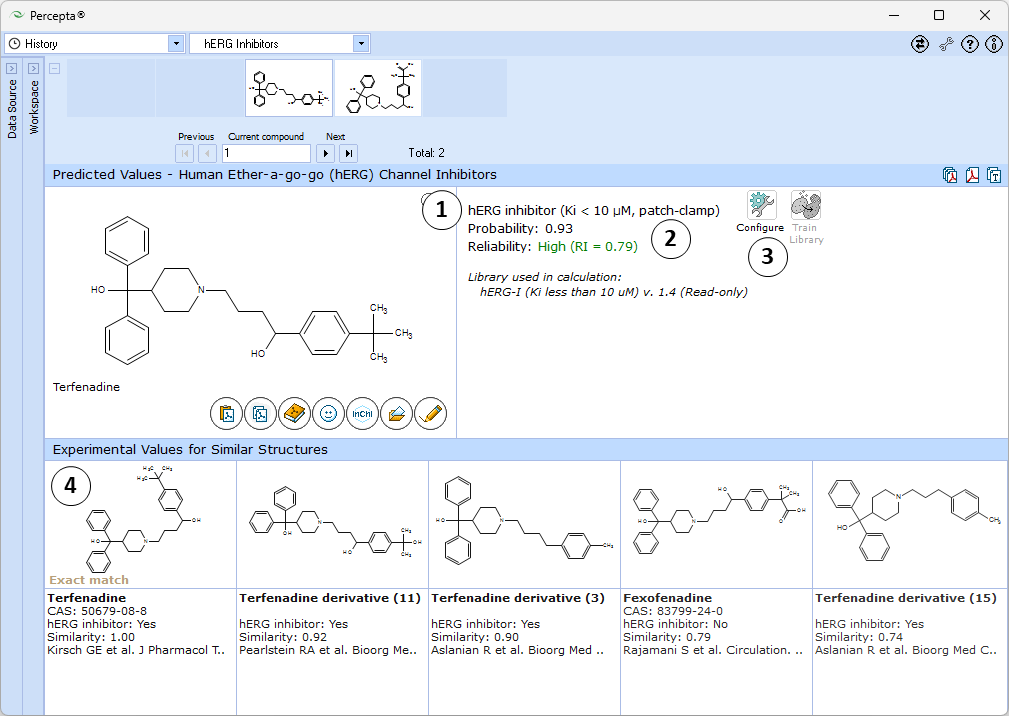
- Estimated probability of the compound being a hERG channel inhibitor.
- Indication of the prediction reliability along with the Reliability Index value:
- RI < 0.3 – Not Reliable,
- RI in range 0.3-0.5 – Borderline Reliability,
- RI in range 0.5-0.75 – Moderate Reliability,
- RI >= 0.75 – High Reliability
- "Configure" and "Train" buttons provide the means to select the training library for use in calculations and to add new data to that library. The names of the currently selected libraries are indicated with italic font
- 5 most similar structures from the self-training library with compound names, experimental classification (Inhibitor or Non-inhibitor), and references
hERG IC50
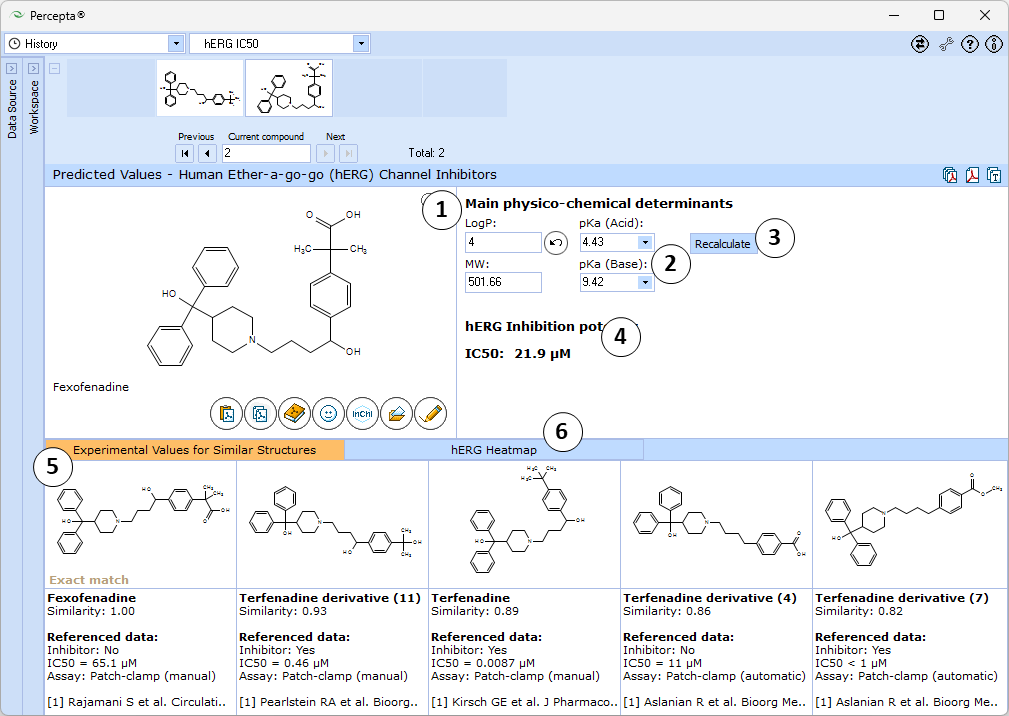
- Main physicochemical descriptors (LogP, Molecular Weight, Acid and Base pKa) that are used as inputs for calculation. By default, automatically calculated values are used. Enter experimental values to improve prediction accuracy or type custom values to model the influence of the respective properties on hERG inhibitory potential of the compound of interest.
- Click the "Undo" button to revert the manually entered property value to the automatically calculated value (LogP and pKa (Acid) in this picture) for a compound and to recalculate hERG inhibition potency
- Click to recalculate hERG inhibition potency using the currently specified parameter values
- Estimated hERG half-inhibitory constant (IC50) of the compound
- 5 most similar structures from the training set with compound names, experimental results (exact IC50 values or intervals), type of assay used for meaurements, and references
- A "heatmap" plot illustrating the partial dependences of hERG inhibition potential on lipophilicity and ionization parameters.
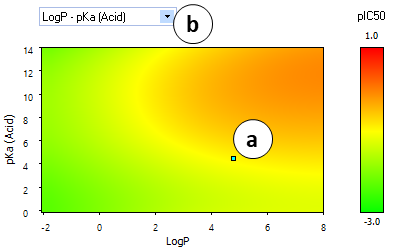
- a. The cyan-colored dot indicates the position of the current compound.
- b. Click to select the variables to plot:
- LogP vs pKa (Acid)
- LogP vs pKa (Base)
- pKa (Acid) vs pKa (Base)
Technical information
Experimental data & predictive models
The current iteration of the built-in library included in hERG inhibition module contains 9383 compounds with experimental values determined using patch-clamp (conventional and automatic) and competitive radioligand displacement assays (reference ligands: dofetilide, astemizole, MK-499). The data were collected from ChEMBL database, as well as original literature publications. Detailed information about the employed data collection and processing procedures can be found in J Comput Aided Mol Des. 2016;30(12):1175-1188. [1]
hERG Inhibition module group contains two different models:
- hERG IC50 module presents a PhysChem-based quantitative model - a Gradient Boosting AFT (Accelerated Failure Time) model that has been trained using both fully quantitative and censored (interval) data. This model predicts predicts IC50 values from a minimal set of physicochemical descriptors including octanol/water LogP, acid and base pKa, molecular size, topology and flexibility parameters. Technical information about the development procedures and performance of this model is available in J Comput Aided Mol Des. 2022;36(12):837-849. [2]
- hERG Inhibitors module provides a probabilistic GALAS model, which is based on a two-part trainable approach, involving a 'baseline' statistical fragmental model, and a similarity-based correction routine that forms the basis of trainability. Originally, this model was built using a smaller data set of 663 molecules with high quality quantitative data. The information provided below applies to that initial model. In the current version of the software, the original 'baseline' model has been trained using the full database of 9383 molecules to ensure the best possible coverage of pharmaceutically relevant chemical space.
Description of the original GALAS model
Data Conversion
The following criteria were applied for conversion of continuous data representing strength of compounds' interaction with hERG channel to binary representation:
- In patch-clamp studies compounds that exhibited IC50 < 10 μM were considered hERG inhibitors, while those with IC50 > 10 μM – hERG non-inhibitors.
- For the data coming from radioligand displacement assay the corresponding thresholds were as follows: Ki < 0.5 μM - inhibitors, Ki > 100 μM - non-inhibitors, while compounds in the intermediate range (0.5 μM < Ki < 100 μM) were labeled inconclusive.
More strict criteria were applied to radioligand displacement data compared to patch-clamp studies since the former method does not provide a direct measure of hERG channel inhibition, but rather represents hERG binding affinity. To ensure high quality of the data set only sufficiently strong or weak binders were considered inhibitors or non-inhibitors respectively, while no definitive categories were assigned to compounds with moderate binding affinities.
Model features & prediction accuracy
Full methodological details of GALAS (Global, Adjusted Locally According to Similarity) modeling approach are available in SAR QSAR Environ Res. 2010;21(1):127-48. [3]
Each GALAS model consists of two parts:
- Global baseline statistical model employing binomial PLS with multiple bootstrapping using a predefined set of fragmental descriptors, that reflects general trends in hERG inhibitory potential.
- Similarity-based routine that performs local correction of baseline predictions considering the differences between baseline and experimental values for the most similar training set compounds.
GALAS methodology also provides the basis for estimating reliability of predictions by the means of calculated Reliability Index (RI) value ranging from 0 to 1 that considers the following two criteria:
- Similarity of tested compound to the training set molecules (prediction is unreliable if no similar compounds have been found).
- Consistence of experimental values and baseline model prediction for the most similar compounds from the training set (discrepant data for similar molecules, i.e. alternating hERG blockers and hERG non-blockers lead to lower RI values).
The used method also provides the basis of model Trainability. 'Trainable model' methodology addresses the issue of the chemical space of ‘in-house’ libraries being considerably wider than that of publicly available data which results in limited applicability of most third-party QSARs for analysis of ‘in-house’ data. The ‘Training engine‘ makes appropriate corrections for systematic deviations produced by the baseline QSAR model based on analysis of similar compounds from the experimental data library. Expansion of this Self-training Library with user-defined experimental data for new compounds leads to instant improvement of prediction accuracy for the respective compound classes. Moreover, addition of 'in-house' data allows adapting the existing model to the particular experimental protocol used in your company and avoiding potential issues related to discrepancies between different experimental methods used for determination of drug interactions with hERG (see Model Trainability Demonstration) section.
The accuracy of predictions for compounds within model Applicability Domain (indicated by Reliability Index values) is comparable to screening results. Predictions that are not reliable, may be instantly improved by addition of experimental data for a few similar compounds to the model Self-training Library.
The table below shows performance of the model on the internal validation set consisting of 151 molecules. Predictions for 103 compounds (68.2% of the validation set) within Model Applicability Domain (indicated by Reliability Index (RI) value > 0.3) are highly accurate:
| Predicted | ||||
|---|---|---|---|---|
| True | False | Accuracy | 91.3% | |
| True | 60 | 4 | Sensitivity | 93.4% |
| False | 5 | 34 | Specificity | 87.2% |
- Only compounds within Applicability Domain (RI > 0.3) were considered in testing.
Model Trainability Demonstration
Trainability of the described predictive model of hERG inhibition was tested using an external data set derived from HTS fluorescence assay that has recently become available in the PubChem database. Validation procedure was performed as follows:
- HTS fluorescence assay data for 1609 compounds were extracted from Pubchem database. Quantitative values provided in the PubChem database (PubChem scores - fluorescence increase over negative control compared to reference compound terfenadine) were converted to binary representation: compounds with Pubchem score > 40% were considered hERG inhibitors; those with Pubchem score from -20 to 20% - non-inhibitors.
- Part of this external data library was reserved as a test set. The remaining data were added to the Self-training Library in three steps.
- The resulting models containing different portions of HTS data were validated against the reserved test set.
When calculations for the test set are made using Built-in Self-training Library, predicted values for many compounds are marked ‘Not reliable’ (i.e. fall outside of the Model Applicability Domain, red bars in the figure). However, as discussed above, prediction accuracy is still impressive if calculations of at least borderline reliability (RI ≥ 0.3) are considered. The key point is the appearance of a considerable number of moderate (RI ≥ 0.5) and high-quality predictions (RI ≥ 0.7) when even a small part of external data set is added to the Self-training Library (green bars in the figure). The percentage of reliable predictions goes even higher with further expansion of the Library, while the same or better overall accuracy of calculations is maintained:
| Reliability | RI > 0.5 | RI > 0.7 | ||
|---|---|---|---|---|
| Library | N | Accuracy | N | Accuracy |
| Built-in | 104 | 96.15% | 6 | 83.33% |
| Built-in + 320 | 302 | 99.01% | 150 | 99.33% |
| Built-in + 623 | 345 | 99.13% | 177 | 99.48% |
| Built-in + 935 | 376 | 98.34% | 192 | 99.48% |
These results demonstrate the ability of our ‘Trainable model’ methodology to adapt the existing model to the chemical space represented by an external compound set. It is also obvious that our Training engine successfully corrects for the differences in experimental estimation when data from different assays are combined and therefore, is particularly suitable for analysis of ‘in-house’ data.