Utilities Menu: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
Enabled pH selector section (v. 2021.2) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[File: | [[File:Spreadsheet_Utilities_Menu_v2021.png]] | ||
<br><br> | <br><br> | ||
Provides several convenient tools for processing the structures of the compounds contained in the spreadsheet: | Provides several convenient tools for processing the structures of the compounds contained in the spreadsheet: | ||
| Line 32: | Line 32: | ||
|- style="vertical-align: top;" | |- style="vertical-align: top;" | ||
|rowspan="2" style="width:250px; text-align:left;" | [[Image:Utilities_menu_check_tautomers.png]] | |rowspan="2" style="width:250px; text-align:left;" | [[Image:Utilities_menu_check_tautomers.png]] | ||
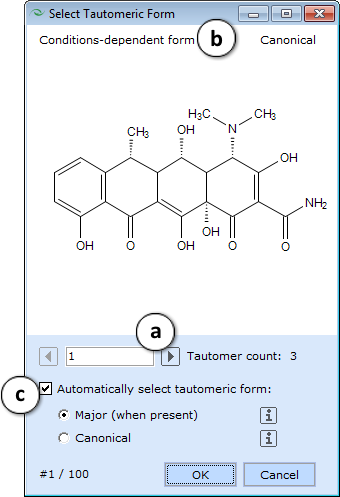
|colspan="2" style="text-align:left;"|'''Check Tautomers''' - Detects possible tautomeric forms of all compounds in the spreadsheet, and for each compound that has more than one form, displays a dialog box that allows selecting which form to keep in the project:<br><br> | |colspan="2" style="text-align:left;"|<span id="Check_tautomers">'''Check Tautomers''' - Detects possible tautomeric forms of all compounds in the spreadsheet, and for each compound that has more than one form, displays a dialog box that allows selecting which form to keep in the project:</span><br><br> | ||
|- style="vertical-align: top; text-align: left;" | |- style="vertical-align: top; text-align: left;" | ||
|[[File:Select Tautomeric Form.png]] | |[[File:Select Tautomeric Form.png]] | ||
| Line 45: | Line 45: | ||
<li>'''Manage Duplicates''' - Checks whether the active project contains any duplicated structures. A dialog box is displayed for specifying structure matching algorithm settings, and how to deal with duplicated records if any are found<br><br>[[File:Utilities_Menu_Manage_Duplicates.png|center]]<br> | <li><span id="Manage_duplicates_window">'''Manage Duplicates'''</span> - Checks whether the active project contains any duplicated structures. A dialog box is displayed for specifying structure matching algorithm settings, and how to deal with duplicated records if any are found<br><br>[[File:Utilities_Menu_Manage_Duplicates.png|center]]<br> | ||
<ol style="list-style-type:lower-latin"> | <ol style="list-style-type:lower-latin"> | ||
<li>The list of options is provided alongside a honeycomb diagram that illustrates whether the central molecule would match (denoted by "=" sign) the molecules on the outside under the currently selected settings.</li> | <li>The list of options is provided alongside a honeycomb diagram that illustrates whether the central molecule would match (denoted by "=" sign) the molecules on the outside under the currently selected settings.</li> | ||
| Line 52: | Line 52: | ||
<li>Check this box to create a new list containing all of the combined entries (type the desired name of the list in the textbox on the right).</li> | <li>Check this box to create a new list containing all of the combined entries (type the desired name of the list in the textbox on the right).</li> | ||
</ol> | </ol> | ||
</li> | |||
<br> | |||
<li><span id="pH_Selector>'''pH Selector'''</span> - Displays the [[pH_Selector]] dialog where you can view LogD curves of the chemical structures from the active project and find the suitable pH range for separation of your mixture. The calculation of LogD is performed considering penetration of ion pairs into organic phase (simulation of physiologic conditions).<br><br><!--[[File:Utilities_Menu_pH_Selector.png|center]]<br>--> | |||
'''Notes:''' | |||
<ul> | |||
<li>pH Selector is only available when you have LogD module licensed in your ACD/Percepta installation</li> | |||
<li>pH selector is restricted to processing at most 100 structures at a time. If there are more than 100 records visible in your active project, only the first 100 will be sent for calculation</li> | |||
<li>LogD calculations in pH selector are always performed using Classic LogP and pKa algorithms regardless of the model settings indicated in the Expert Panel</li> | |||
</ul> | |||
</li> | </li> | ||
</ol> | </ol> | ||
Latest revision as of 13:08, 31 January 2022
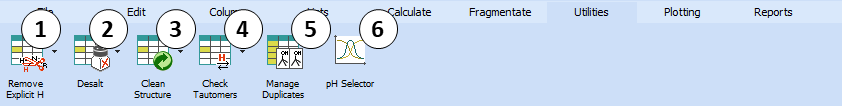
Provides several convenient tools for processing the structures of the compounds contained in the spreadsheet:
-
Remove Explicit H - Removes all explicitly drawn hydrogen atoms (converts them to the implicit form). 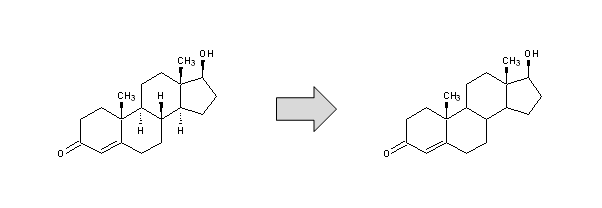
-
Desalt - If the structure corresponds to a salt, removes the smaller counterion, and if possible, converts the remaining ion into a neutral form. 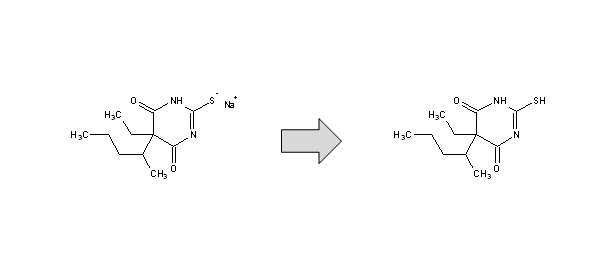
-
Clean Structure - Automatically re-assigns 2D coordinates of atoms in order to provide the drawn structure a more visually appealing ("clean") look. 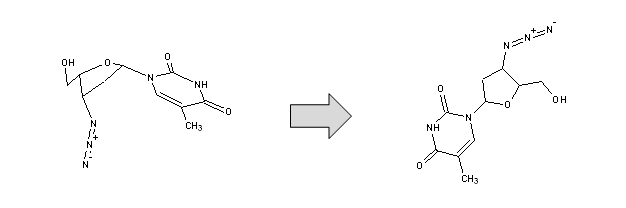
- Manage Duplicates - Checks whether the active project contains any duplicated structures. A dialog box is displayed for specifying structure matching algorithm settings, and how to deal with duplicated records if any are found
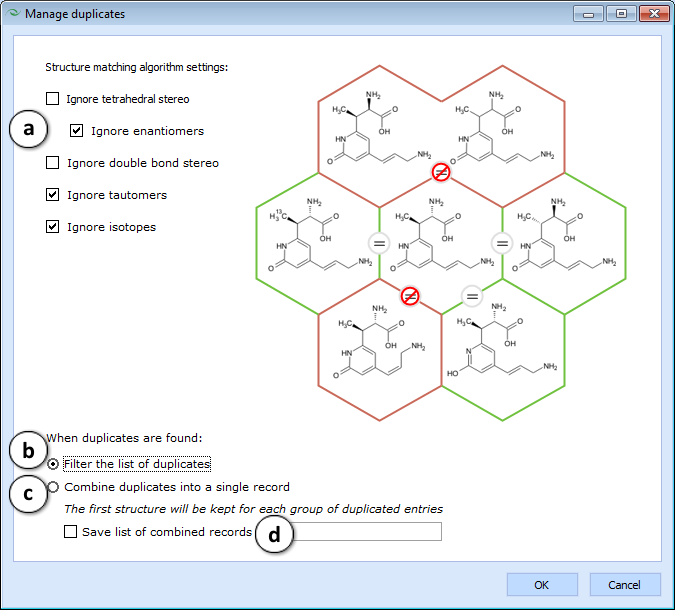
- The list of options is provided alongside a honeycomb diagram that illustrates whether the central molecule would match (denoted by "=" sign) the molecules on the outside under the currently selected settings.
- If this option is selected, the program will not take any action with respect to found duplicates, but will only filter the spreadsheet so that only duplicated records are visible, and sorted in such way, that the matching structures will be displayed alongside each other.
- If this option is selected, the program will keep only the first of each group of matching structures in the project, while any numerical or text data associated with duplicated records will be combined into cells with Multiple Entries.
- Check this box to create a new list containing all of the combined entries (type the desired name of the list in the textbox on the right).
- pH Selector - Displays the pH_Selector dialog where you can view LogD curves of the chemical structures from the active project and find the suitable pH range for separation of your mixture. The calculation of LogD is performed considering penetration of ion pairs into organic phase (simulation of physiologic conditions).
Notes:- pH Selector is only available when you have LogD module licensed in your ACD/Percepta installation
- pH selector is restricted to processing at most 100 structures at a time. If there are more than 100 records visible in your active project, only the first 100 will be sent for calculation
- LogD calculations in pH selector are always performed using Classic LogP and pKa algorithms regardless of the model settings indicated in the Expert Panel