ADMET Profiler: Difference between revisions
Added technical section with categorization function info |
m Renamed module in the program |
||
| (4 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Overview== | ==Overview== | ||
''' | '''ADMET Profiler''' is an integrated workspace that aggregates the output of various PhysChem, ADME, and Drug safety-related predictors included in '''ACD/Percepta''' and presents the data as the compound's overall ADME/Tox profile with individual predictions categorized according to user-adjustable thresholds and visualized using color-coded indications. ADMET Profiler enables the researcher to identify the most suitable structures that meet specific criteria for a wide range of properties: | ||
* PhysChem | * PhysChem | ||
** LogP | ** LogP | ||
| Line 11: | Line 11: | ||
** Human Intestinal Absorption (HIA) and Caco-2 permeability | ** Human Intestinal Absorption (HIA) and Caco-2 permeability | ||
** Plasma protein binding (PPB) | ** Plasma protein binding (PPB) | ||
** Blood-brain | ** Blood-brain penetration and accessibility to CNS | ||
** Metabolic stability | ** Metabolic stability | ||
* Drug Safety | * Drug Safety | ||
| Line 24: | Line 24: | ||
<br> | <br> | ||
<ol> | <ol> | ||
<li>'''Navigation Bar'''<br> The '''Navigation Bar''' appears in all workspaces except the '''Spreadsheet View'''. It displays presented structure and two previous as well as two next | <li>'''Navigation Bar'''<br> The '''Navigation Bar''' appears in all workspaces except the '''Spreadsheet View'''. It displays presented structure and two previous as well as two next structures in the corresponding dataset. If no dataset is selected, the '''Navigation Bar''' displays the history of performed calculations. <br> When the compound outside of the dataset is used as an input structure for calculation, the user can add it to the current dataset:<br><br> [[File:Navigation_bar.png|center]]<br> | ||
<ol style="list-style-type:lower-latin"> | <ol style="list-style-type:lower-latin"> | ||
<li>Click the [[File:History_confirm_icon.png]] pictogram to replace the current structure with the new one. All subsequent calculations will be performed automatically for the added compound. </li> | <li>Click the [[File:History_confirm_icon.png]] pictogram to replace the current structure with the new one. All subsequent calculations will be performed automatically for the added compound. </li> | ||
| Line 34: | Line 34: | ||
<li>'''Structure Pane'''<br>The '''Structure Pane''' displays a 2-D representation of the chemical structure used for calculation. A compound name is displayed only for those structures which are found in the '''Dictionary'''.<br>[[Image:Paste.png]] '''(Ctrl+V)''': Pastes an input structure from the clipboard to the '''Structure Pane'''. Also, if you are pasting a SMILES string from the clipboard, you can save a step by clicking directly on this button to display the 2-D structure. The calculation will be performed automatically for the input structure.<br>[[Image:Copy_pictogram.png]] '''(Ctrl+C)''': Copies the active structure in the '''Structure Pane''' to the clipboard.<br>[[Image:Opendictionary.png]] '''(Ctrl+D)''': Opens the '''Dictionary''' with >117,000 available names to find the corresponding structure and display it. When a compound is selected from the list and the selection confirmed by clicking '''OK''', calculations for the chosen compound will be performed automatically.<br>[[Image:SMILES_pictogram.png]] '''(Ctrl+S)''': Type or paste SMILES notation string in this field. The string will automatically be converted to 2-D structure. Calculation will be performed automatically for the input structure.<br> [[Image:Open_pictogram.png]] '''(Ctrl+O)''': Opens a file containing a molecular structure. '''ACD/Percepta''' supports all versions of MDL *.mol files, ACD/ChemSketch *.sk2, as well as ISIS Draw *.skc, and ChemDraw *.cdx formats.<br>[[Image:Edit_pictogram.png]] '''(Ctrl+E)''': Allows to edit the active molecule. </li><br> | <li>'''Structure Pane'''<br>The '''Structure Pane''' displays a 2-D representation of the chemical structure used for calculation. A compound name is displayed only for those structures which are found in the '''Dictionary'''.<br>[[Image:Paste.png]] '''(Ctrl+V)''': Pastes an input structure from the clipboard to the '''Structure Pane'''. Also, if you are pasting a SMILES string from the clipboard, you can save a step by clicking directly on this button to display the 2-D structure. The calculation will be performed automatically for the input structure.<br>[[Image:Copy_pictogram.png]] '''(Ctrl+C)''': Copies the active structure in the '''Structure Pane''' to the clipboard.<br>[[Image:Opendictionary.png]] '''(Ctrl+D)''': Opens the '''Dictionary''' with >117,000 available names to find the corresponding structure and display it. When a compound is selected from the list and the selection confirmed by clicking '''OK''', calculations for the chosen compound will be performed automatically.<br>[[Image:SMILES_pictogram.png]] '''(Ctrl+S)''': Type or paste SMILES notation string in this field. The string will automatically be converted to 2-D structure. Calculation will be performed automatically for the input structure.<br> [[Image:Open_pictogram.png]] '''(Ctrl+O)''': Opens a file containing a molecular structure. '''ACD/Percepta''' supports all versions of MDL *.mol files, ACD/ChemSketch *.sk2, as well as ISIS Draw *.skc, and ChemDraw *.cdx formats.<br>[[Image:Edit_pictogram.png]] '''(Ctrl+E)''': Allows to edit the active molecule. </li><br> | ||
<li>[[Image:Create_pdf_pictogram.png]] Creates a PDF report with the displayed results.<br>[[Image:Copy_results_pictogram.png]] Copies the displayed results to the clipboard.</li><br> | <li>[[Image:Batch_pdf_pictogram.png]] Saves a PDF report for each compound in the active project in the directory of your choice.<br>[[Image:Create_pdf_pictogram.png]] Creates a PDF report with the displayed results.<br>[[Image:Copy_results_pictogram.png]] Copies the displayed results to the clipboard.</li><br> | ||
<li>The dropdown list of available threshold templates. Use default thresholds or select any of the previously saved templates from the dropdown, and the property categories will be automatically re-assigned to match the newly selected set of thresholds. If any of the thresholds are altered compared to last saved version, this is indicated by an asterisk displayed next to the name of the template. The changes can be saved under the same template name, by pressing '''Save''' button, or alternatively, | <li>The dropdown list of available threshold templates. Use default thresholds or select any of the previously saved templates from the dropdown, and the property categories will be automatically re-assigned to match the newly selected set of thresholds. If any of the thresholds are altered compared to last saved version, this is indicated by an asterisk displayed next to the name of the template. The changes can be saved under the same template name, by pressing '''Save''' button, or alternatively, transferred to a new template by choosing '''Save as...''' command.</li><br> | ||
<li>A list of properties included in the profile, their calculated values, and category indication with a short textual description. The property list is interactive - click the property of interest to display its value scale and view or edit the cut-offs separating the categories in the lower left part of the window.</li><br> | <li>A list of properties included in the profile, their calculated values, and category indication with a short textual description. The property list is interactive - click the property of interest to display its value scale and view or edit the cut-offs separating the categories in the lower left part of the window.</li><br> | ||
| Line 45: | Line 45: | ||
</ol> | </ol> | ||
<div class="mw-collapsible | <div class="mw-collapsible"> | ||
==Technical information== | ==Technical information== | ||
| Line 51: | Line 51: | ||
<div class="mw-collapsible-content"> | <div class="mw-collapsible-content"> | ||
All categorization functions in | All categorization functions in ADMET Profiler can be divided into two large groups: | ||
* Based on continuous properties | * Based on continuous properties | ||
* Based on probabilistic predictors | * Based on probabilistic predictors | ||
Latest revision as of 08:21, 14 August 2024
Overview
ADMET Profiler is an integrated workspace that aggregates the output of various PhysChem, ADME, and Drug safety-related predictors included in ACD/Percepta and presents the data as the compound's overall ADME/Tox profile with individual predictions categorized according to user-adjustable thresholds and visualized using color-coded indications. ADMET Profiler enables the researcher to identify the most suitable structures that meet specific criteria for a wide range of properties:
- PhysChem
- LogP
- Solubility
- H-bonding
- Molecular size and flexibility
- Drug- and Lead-likeness rules
- ADME
- Human Intestinal Absorption (HIA) and Caco-2 permeability
- Plasma protein binding (PPB)
- Blood-brain penetration and accessibility to CNS
- Metabolic stability
- Drug Safety
- P-gp substrate specificity
- Cytochrome P450 inhibitor specificity
- hERG inhibitor specificity
- Mutagenicity in Ames test
Interface
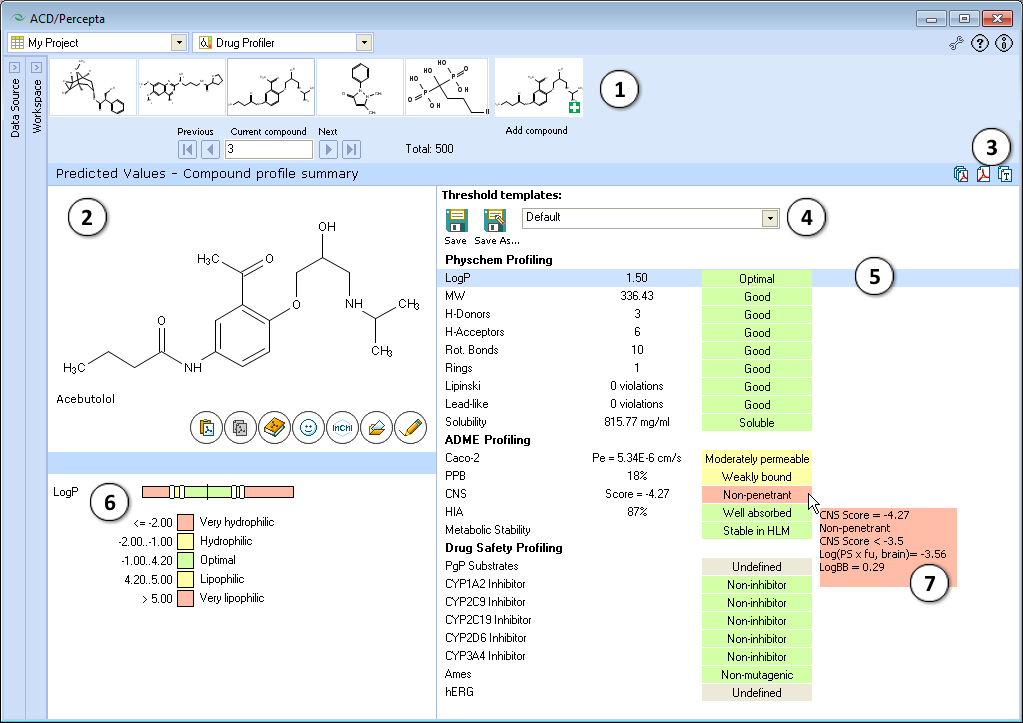
- Navigation Bar
The Navigation Bar appears in all workspaces except the Spreadsheet View. It displays presented structure and two previous as well as two next structures in the corresponding dataset. If no dataset is selected, the Navigation Bar displays the history of performed calculations.
When the compound outside of the dataset is used as an input structure for calculation, the user can add it to the current dataset:
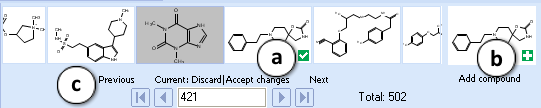
- Click the
 pictogram to replace the current structure with the new one. All subsequent calculations will be performed automatically for the added compound.
pictogram to replace the current structure with the new one. All subsequent calculations will be performed automatically for the added compound. - Click the
 pictogram to add new compound to the current dataset. It will appear as the last record in the dataset and all subsequent calculations will be performed automatically.
pictogram to add new compound to the current dataset. It will appear as the last record in the dataset and all subsequent calculations will be performed automatically. - The structure browse bar displays the active structure number and the total number of records in the current dataset. Scroll through the database using the browse buttons: First/Last record, Previous/Next record or pressing the up/down arrow keys on the keyboard to go to the next/previous record. Type a record number to view it directly.
- Click the
- Structure Pane
The Structure Pane displays a 2-D representation of the chemical structure used for calculation. A compound name is displayed only for those structures which are found in the Dictionary. (Ctrl+V): Pastes an input structure from the clipboard to the Structure Pane. Also, if you are pasting a SMILES string from the clipboard, you can save a step by clicking directly on this button to display the 2-D structure. The calculation will be performed automatically for the input structure.
(Ctrl+V): Pastes an input structure from the clipboard to the Structure Pane. Also, if you are pasting a SMILES string from the clipboard, you can save a step by clicking directly on this button to display the 2-D structure. The calculation will be performed automatically for the input structure. (Ctrl+C): Copies the active structure in the Structure Pane to the clipboard.
(Ctrl+C): Copies the active structure in the Structure Pane to the clipboard. (Ctrl+D): Opens the Dictionary with >117,000 available names to find the corresponding structure and display it. When a compound is selected from the list and the selection confirmed by clicking OK, calculations for the chosen compound will be performed automatically.
(Ctrl+D): Opens the Dictionary with >117,000 available names to find the corresponding structure and display it. When a compound is selected from the list and the selection confirmed by clicking OK, calculations for the chosen compound will be performed automatically. (Ctrl+S): Type or paste SMILES notation string in this field. The string will automatically be converted to 2-D structure. Calculation will be performed automatically for the input structure.
(Ctrl+S): Type or paste SMILES notation string in this field. The string will automatically be converted to 2-D structure. Calculation will be performed automatically for the input structure.
 (Ctrl+O): Opens a file containing a molecular structure. ACD/Percepta supports all versions of MDL *.mol files, ACD/ChemSketch *.sk2, as well as ISIS Draw *.skc, and ChemDraw *.cdx formats.
(Ctrl+O): Opens a file containing a molecular structure. ACD/Percepta supports all versions of MDL *.mol files, ACD/ChemSketch *.sk2, as well as ISIS Draw *.skc, and ChemDraw *.cdx formats. (Ctrl+E): Allows to edit the active molecule.
(Ctrl+E): Allows to edit the active molecule.  Saves a PDF report for each compound in the active project in the directory of your choice.
Saves a PDF report for each compound in the active project in the directory of your choice. Creates a PDF report with the displayed results.
Creates a PDF report with the displayed results. Copies the displayed results to the clipboard.
Copies the displayed results to the clipboard.- The dropdown list of available threshold templates. Use default thresholds or select any of the previously saved templates from the dropdown, and the property categories will be automatically re-assigned to match the newly selected set of thresholds. If any of the thresholds are altered compared to last saved version, this is indicated by an asterisk displayed next to the name of the template. The changes can be saved under the same template name, by pressing Save button, or alternatively, transferred to a new template by choosing Save as... command.
- A list of properties included in the profile, their calculated values, and category indication with a short textual description. The property list is interactive - click the property of interest to display its value scale and view or edit the cut-offs separating the categories in the lower left part of the window.
- Displays the value scale of corresponding property and the list of cut-offs defining different categories. A black vertical line indicates the position of the current compound on the property scale. Move the sliders to alter the numerical cut-offs separating the categories.
- Hover over the property value to view a screen-tip with more details regarding the particular prediction. For some properties, the screen-tips display additional quantitative parameters of relevance (e.g., CNS), predicted probabilities of positive assay outcome in case of probabilistic properties, and/or the numerical conditions that the compound had fulfilled in obtain to be classified in a particular category.
Technical information
All categorization functions in ADMET Profiler can be divided into two large groups:
- Based on continuous properties
- Based on probabilistic predictors
Continuous properties
For continuous properties the categories are defined by setting-up certain numerical thresholds on the scale of the corresponding property values. The following is a complete list of default thresholds along with additional comments where appropriate.
PhysChem Profiling
The majority of the PhysChem related categorization functions employ upper property value thresholds introduced by Christopher A. Lipinski in his "Rule of 5". In some cases additional cut-offs on the lower end of the scale have been introduced as well as certain intermediate ranges resulting in the Average outcome. The latter are mostly associated with the concept of lead-likeness which is described later in this text.
Lipophilicity
| Value range | Category |
|---|---|
| logP <= -2 | Very hydrophilic |
| -2 < logP <= -1 | Hydrophilic |
| -1 < logP <= 4.2 | Optimal |
| 4.2 < logP <= 5 | Hydrophobic |
| logP > 5 | Very hydrophobic |
Molecular weight
| Value range | Category |
|---|---|
| MW <= 460 | Good |
| 460 < MW <= 500 | Moderate |
| MW > 500 | Bad |
Hydrogen bonding acidity
| Value range | Category |
|---|---|
| No. of H-Bond donors <= 5 | Good |
| No. of H-Bond donors > 5 | Bad |
Hydrogen bonding basicity
| Value range | Category |
|---|---|
| No. of H-Bond acceptors <= 10 | Good |
| No. of H-Bond acceptors > 10 | Bad |
"Rule of 5"
The Lipinski categorization function acts as a sort of a summary of the above separate categories, by testing a compound against all rules at once (-2 =< logP < 5; MW < 500; No. of H-Bond donors <= 5; No. of H-Bond acceptors <= 10). Category assignments are the following:
| No. of rule violations | Category |
|---|---|
| 0 (All 4 rules apply) | Good |
| 1..2 (Any 2 or 3 rules apply) | Moderate |
| >2 (Less than 2 rules apply) | Bad |
Lead-likeness
The Lead-like category assignment functions in exactly the same way, however a little bit lower property value thresholds are usually employed. This concept is based on the assumption that the lead compound undergoes various modifications during the development stage and establishment of certain trends in which way those modifications actually affect the physicochemical properties of the lead compound. In other words, it is an attempt to establish the rules for filtering out the compounds that are unlikely to violate Lipinski’s "Rule of 5" after the development stage (-2 =< logP < 4.2; MW < 460; No. of H-Bond donors <= 5; No. of H-Bond acceptors <= 9).
There are also a few additional categorization functions in the PhysChem Profiling group. Flexibility of the molecule
| Value range | Category |
|---|---|
| No. of rotatable bonds <= 10 | Good |
| No. of rotatable bonds > 10 | Bad |
Number of Rings
| Value range | Category |
|---|---|
| No. of Rings <= 4 | Good |
| No. of Rings > 4 | Bad |
Solubility
The classification scheme in case of the compound solubility is a slightly modified version of the Traffic Lights system published in Lobell, M., et al. Chem. Med. Chem., 2006, 1, 1229-1236. It takes into account the solubility of the compound in buffer at pH of 6.5.
| Solubility at pH=6.5 | Category |
|---|---|
| S > 100 mg/L | Soluble |
| 10 mg/L < S <= 100 mg/L | Insoluble |
| S <= 10 mg/L | Highly insoluble |
ADME Profiling
Plasma protein binding
Categorization function for the extent of plasma protein binding takes into account both predicted %PPB values and Reliability Index (RI) of predictions:
| Value range | Category |
|---|---|
| Any value with RI < 0.3 | Undefined |
| %PPB <= 10% | Not bound |
| 10% < %PPB <= 40% | Weakly bound |
| 40% < %PPB <= 80% | Moderately bound |
| 80% < %PPB <= 90% | Strongly bound |
| %PPB > 90% | Extensively bound |
Caco-2 Permeability
Category assignment is based on predicted permeability across Caco-2 monolayers (Pe) at pH = 7.4 and 500 rpm stirring rate:
| Value range | Category |
|---|---|
| Pe <= 1 * 10-6 cm/s | Poorly permeable |
| 1 * 10-6 cm/s < Pe <= 7 * 10-6 cm/s | Moderately permeable |
| Pe > 7 * 10-6 cm/s | Highly permeable |
Human Intestinal Absorption
| Value range | Category |
|---|---|
| %HIA <= 30% | Poorly permeable |
| 30% < %HIA <= 70% | Moderately permeable |
| %HIA > 70% | Highly permeable |
CNS access
Evaluation of compounds’ brain penetration potential and their classification by CNS access is based on predicted quantitative characteristics of transport across BBB, namely brain/plasma equilibration rate, Log (PS x fu,brain) and steady-state brain/plasma distribution ratio, LogBB. The categories are assigned according to the values of CNS Access Score which is calculated as a simple combination of those parameters: Score = Log(PS x fu,brain) + LogBB.
| Score range | Category | Comment |
|---|---|---|
| Score <= -3.5 | Non-penetrant | Brain delivery too low for a compound to be active in CNS |
| -3.5 < Score <= -3 | Weak penetrant | Borderline brain delivery potential |
| Score > -3 | Penetrant | Compound sufficiently accessible to CNS to exhibit pharmacological effect in brain |
The above mentioned parameters reflect compounds' transport across BBB governed by passive diffusion only. Therefore, CNS Access Score may also be shifted up or down if the respective compound is predicted to undergo carrier-mediated transport at the BBB. The Score is increased by 0.5 units for probable substrates of influx transporters, and decreased by 1 unit for compounds potentially susceptible to P-gp efflux. These are the final Score values used in category assignment.
Probabilistic categories
In probabilistic categorization functions the numerical predictions are first converted to the so called classification Scores on the basis of the obtained probability (p) and Reliability Index (RI) values. The conversion is performed according to the following equation: Score = (p - 0.5) * RI + 0.5
The main idea lying behind this transformation is that predicted probability close to 0.5 and low RI are both indicators of an inconclusive prediction. Therefore, the amount by which the initially calculated probability differs from 0.5 is multiplied by RI, so that predictions with low RI values ultimately yield scores in the intermediate range even if the original p-value is quite high or low. The final category assignment takes place on the basis of the defined classification score ranges.
This class of probabilistic categorization functions covers the Metabolic Stability category in the ADME Profiling group and the entire Drug Safety Profiling group:
- Ames categories
- hERG categories
- Pgp Substrates categories
- CYP450 Inhibitor categories (3A4, 2D6, 2C9, 2C19, 1A2)
In case of AMES, hERG and Metabolic Stability categories everything applies as explained. Since the classification criteria for all of these properties are such that a positive outcome is considered unfavorable for a drug-candidate (i.e., Ames positive, hERG inhibitor with IC50 < 10 uM, and t1/2 in Human Liver Microsomes < 30 min., respectively), high probabilities result in a classification as Bad, i.e.:
| Score range | Category label | ||
|---|---|---|---|
| Ames | hERG | HLM | |
| Score <= 0.33 | Non-mutagenic | Non-inhibitor | Stable in HLM |
| 0.33 < Score <= 0.67 | Undefined | Undefined | Undefined |
| Score > 0.67 | Mutagenic | Inhibitor | Unstable in HLM |
P-gp Substrate and Cytochrome P450 inhibition categories constitute special cases as they are assigned on the basis of two predictions each. In case of P-gp:
- Score1 is derived from Good P-gp substrate model (compounds are only considered positive if they undergo significant P-gp efflux in vivo)
- Score2 is derived from General P-gp substrate model
In case of all considered cytochrome P450 isoforms, the two models correspond to different IC50 thresholds for classification:
- Score1 - Efficient inhibition model (IC50 < 10 uM)
- Score2 - General inhibition model (IC50 < 50 uM)
The categories are assigned as follows:
| Score1 | Score2 | Category label | |
|---|---|---|---|
| P-gp | CYP450 | ||
| Score1 > 0.67 | Score2 - not relevant | Good substrate | Efficient inhibitor |
| Score1 <= 0.67 | Score2 > 0.67 | Substrate | Inhibitor |
| Score1 <= 0.67 | 0.33 < Score2 <= 0.67 | Undefined | Undefined |
| Score1 <= 0.67 | Score2 <= 0.33 | Non-substrate | Non-inhibitor |