Trainable Libraries: Difference between revisions
Created page with "As a starting point for the calculations a number of Built-in Self-training Libraries with experimental values of the corresponding properties is provided for each ‘Trainabl..." |
mNo edit summary |
||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 18: | Line 18: | ||
** %PPB v. 1.2 - 1453 compounds. | ** %PPB v. 1.2 - 1453 compounds. | ||
* '''Trainable LogP''' | * '''Trainable LogP''' | ||
** LogP v. 1. | ** LogP v. 1.3 - 17854 compounds. | ||
* '''Trainable LogD''' | * '''Trainable LogD''' | ||
** Training takes place through LogP Self-training Libraries. The training procedure implemented in ACD/LogP GALAS module accepts LogD values measured at any pH and automatically recalculates these to the respective LogP of neutral species to be stored in the library. The trained LogP library may then be used in both LogP and LogD calculations. | ** Training takes place through LogP Self-training Libraries. The training procedure implemented in ACD/LogP GALAS module accepts LogD values measured at any pH and automatically recalculates these to the respective LogP of neutral species to be stored in the library. The trained LogP library may then be used in both LogP and LogD calculations. | ||
| Line 54: | Line 54: | ||
* '''Trainable LC50 P. promelas''' | * '''Trainable LC50 P. promelas''' | ||
** LC50 P. promelas v. 1.2 - 900 compounds. | ** LC50 P. promelas v. 1.2 - 900 compounds. | ||
* '''Trainable IGC50 T. pyriformis''' | |||
** IGC50 T. pyriformis v. 1.0 - 1087 compounds. | |||
* '''Trainable LD50 Mouse IP''' | * '''Trainable LD50 Mouse IP''' | ||
** LD50 Mouse Intraperitoneal v. 1.2 - 36030 compounds. | ** LD50 Mouse Intraperitoneal v. 1.2 - 36030 compounds. | ||
| Line 67: | Line 69: | ||
** LD50 Rat Oral v. 1.2 - 8631 compounds. | ** LD50 Rat Oral v. 1.2 - 8631 compounds. | ||
* '''Trainable hERG I''' | * '''Trainable hERG I''' | ||
** hERG-I (Ki less than 10 uM) - | ** hERG-I (Ki less than 10 uM) v. 1.4 - 9383 compounds. | ||
* '''Trainable Ames''' | * '''Trainable Ames''' | ||
** AMES Test v. 1. | ** AMES Test v. 1.3 - 10294 compounds. | ||
<br /> | <br /> | ||
<!--'''Note''': The size of ''Built-in pKa Self-training Library'' is given not as a number of compounds, but rather as a total number of entries, since experimental data for several ionogenic centers in the same molecule may be present in the library.<br /> | <!--'''Note''': The size of ''Built-in pKa Self-training Library'' is given not as a number of compounds, but rather as a total number of entries, since experimental data for several ionogenic centers in the same molecule may be present in the library.<br /> | ||
<br />--> | <br />--> | ||
All built-in libraries are ‘Read-only’ meaning that the user cannot alter their contents. In order to supplement built-in data with user-defined data, both read-only and user created libraries can be used in calculations together. To change any values contained in the built-in library, one can create a new library, and copy the contents of the built-in library using Import > Import > Import directly from trainable libraries command.<br> | |||
<br> | |||
<br> | |||
[[File:trainable_libraries_create_new.png|right]]<br>In order to create a new library, click “New Library” in the appropriate '''Trainable Libraries''' group. In the '''Create New Self-training Library''' window, type in:<br> | |||
:a. Library name<br> | |||
:b. Library description.<br> | |||
<br> | <br> | ||
Browse for a folder where you wish to save the new library, and enter the desired name of the *.trn file. The newly added library appears under the respective '''Trainable Libraries''' group and is automatically loaded in the '''Spreadsheet''' workspace.<br> | Browse for a folder where you wish to save the new library, and enter the desired name of the *.trn file. The newly added library appears under the respective '''Trainable Libraries''' group and is automatically loaded in the '''Spreadsheet''' workspace.<br> | ||
<br> | <br> | ||
Click “Add Existing” in the appropriate '''Trainable Libraries''' group to browse for an existing training library in TRN format. If the selected *.trn file belongs to another group, the program displays a warning, and after clicking OK automatically reallocates the library to the correct group.<br> | |||
<br> | <br> | ||
Right-click the user-defined training library to remove it from the list or to rename.<br> | Right-click the user-defined training library to remove it from the list or to rename.<br> | ||
[[File:trainable_libraries_rightclick.png]] | [[File:trainable_libraries_rightclick.png]]<br> | ||
<br> | |||
<br> | <br> | ||
''For more information see [[Trainable Models]] and [[Training]]''<br> | ''For more information see [[Trainable Models]] and [[Training]]''<br> | ||
Latest revision as of 10:39, 26 July 2023
As a starting point for the calculations a number of Built-in Self-training Libraries with experimental values of the corresponding properties is provided for each ‘Trainable Model’ in ACD/Percepta:
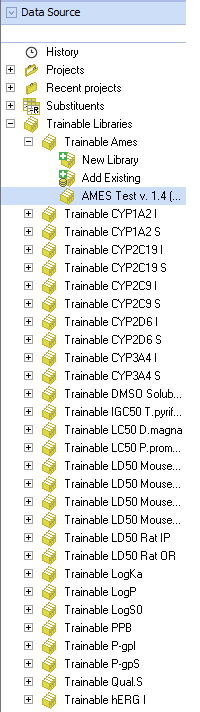
- Trainable P-gpS
- P-gpS v. 1.2 - 1596 compounds.
- Trainable P-gpI
- P-gpI v. 1.2 - 2006 compounds.
- Trainable LogS0
- LogS0 v. 1.2 - 6806 compounds.
- Trainable LogS
- Training takes place through LogS0 Self-training Libraries. The training procedure implemented in ACD/LogS0 GALAS module accepts both LogSw measured in pure water and LogS at any pH. These values are automatically recalculated to the respective LogS0 of neutral form to be stored in the library. The trained LogS0 library may then be used for LogS0, LogSw, and LogS calculations.
- Trainable LogKa
- LogKa(HSA) v. 1.2 - 334 compounds.
- Trainable PPB
- %PPB v. 1.2 - 1453 compounds.
- Trainable LogP
- LogP v. 1.3 - 17854 compounds.
- Trainable LogD
- Training takes place through LogP Self-training Libraries. The training procedure implemented in ACD/LogP GALAS module accepts LogD values measured at any pH and automatically recalculates these to the respective LogP of neutral species to be stored in the library. The trained LogP library may then be used in both LogP and LogD calculations.
- Trainable CYP1A2 I
- CYP1A2-I (IC50 less than 10 uM) v. 1.2 - 5815 compounds.
- CYP1A2-I (IC50 less than 50 uM) v. 1.2 - 4867 compounds.
- Trainable CYP2C19 I
- CYP2C19-I (IC50 less than 10 uM) v. 1.2 - 6833 compounds.
- CYP2C19-I (IC50 less than 50 uM) v. 1.2 - 6899 compounds.
- Trainable CYP2C9 I
- CYP2C9-I (IC50 less than 10 uM) v. 1.2 - 7677 compounds.
- CYP2C9-I (IC50 less than 50 uM) v. 1.2 - 7666 compounds.
- Trainable CYP2D6 I
- CYP2D6-I (IC50 less than 10 uM) v. 1.2 - 7507 compounds.
- CYP2D6-I (IC50 less than 50 uM) v. 1.2 - 7707 compounds.
- Trainable CYP3A4 I
- CYP3A4-I (IC50 less than 10 uM) v. 1.2 - 7926 compounds.
- CYP3A4-I (IC50 less than 50 uM) v. 1.2 - 6683 compounds.
- Trainable CYP1A2 S
- CYP1A2-S v. 1.2 - 935 compounds.
- Trainable CYP2C19 S
- CYP2C19-S v. 1.2 - 794 compounds.
- Trainable CYP2C9 S
- CYP2C9-S v. 1.2 - 867 compounds.
- Trainable CYP2D6 S
- CYP2D6-S v. 1.2 - 1001 compounds.
- Trainable CYP3A4 S
- CYP3A4-S v. 1.2 - 960 compounds.
- Trainable DMSO Solubility
- S(DMSO) > 20 mM v. 1.2 - 22262 compounds.
- Trainable LC50 D. magna
- LC50 D. magna v. 1.2 - 588 compounds.
- Trainable LC50 P. promelas
- LC50 P. promelas v. 1.2 - 900 compounds.
- Trainable IGC50 T. pyriformis
- IGC50 T. pyriformis v. 1.0 - 1087 compounds.
- Trainable LD50 Mouse IP
- LD50 Mouse Intraperitoneal v. 1.2 - 36030 compounds.
- Trainable LD50 Mouse IV
- LD50 Mouse Intravenous v. 1.2 - 19961 compounds.
- Trainable LD50 Mouse OR
- LD50 Mouse Oral v. 1.2 - 19569 compounds.
- Trainable LD50 Mouse SC
- LD50 Mouse Subcutaneous v. 1.2 - 8575 compounds.
- Trainable LD50 Rat IP
- LD50 Rat Intraperitoneal v. 1.2 - 5002 compounds.
- Trainable LD50 Rat OR
- LD50 Rat Oral v. 1.2 - 8631 compounds.
- Trainable hERG I
- hERG-I (Ki less than 10 uM) v. 1.4 - 9383 compounds.
- Trainable Ames
- AMES Test v. 1.3 - 10294 compounds.
All built-in libraries are ‘Read-only’ meaning that the user cannot alter their contents. In order to supplement built-in data with user-defined data, both read-only and user created libraries can be used in calculations together. To change any values contained in the built-in library, one can create a new library, and copy the contents of the built-in library using Import > Import > Import directly from trainable libraries command.
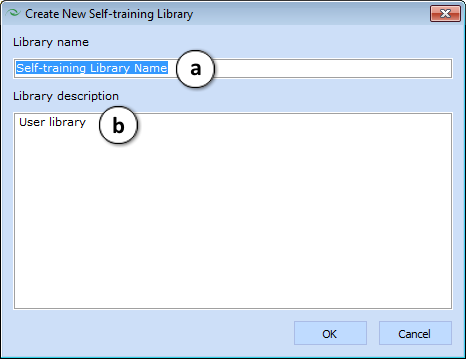
In order to create a new library, click “New Library” in the appropriate Trainable Libraries group. In the Create New Self-training Library window, type in:
- a. Library name
- b. Library description.
Browse for a folder where you wish to save the new library, and enter the desired name of the *.trn file. The newly added library appears under the respective Trainable Libraries group and is automatically loaded in the Spreadsheet workspace.
Click “Add Existing” in the appropriate Trainable Libraries group to browse for an existing training library in TRN format. If the selected *.trn file belongs to another group, the program displays a warning, and after clicking OK automatically reallocates the library to the correct group.
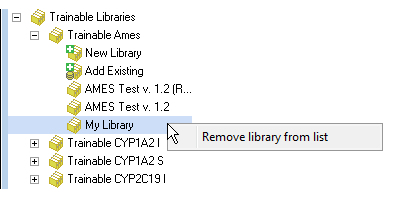
Right-click the user-defined training library to remove it from the list or to rename.
For more information see Trainable Models and Training