Absorption Caco2: Difference between revisions
| Line 6: | Line 6: | ||
===Features=== | ===Features=== | ||
* Calculates the extent of passive permeability of analyzed compounds across | * Calculates the extent of passive permeability of analyzed compounds across Caco-2 cell monolayers at specified pH and stirring conditions using physicochemical property values such as lipophilicity (LogP) and ionization (pKa) as inputs. | ||
* Gives an estimate of relative contributions of different transport routes | * Gives an estimate of relative contributions of different transport routes to Caco-2 permeability. | ||
* Allows entering experimentally measured physicochemical properties instead of automatically calculated values to improve the quality of predictions. | * Allows entering experimentally measured physicochemical properties instead of automatically calculated values to improve the quality of predictions. The experimental conditions (pH and stirring rate) may also be adjusted manually for full flexibility of the simulation. | ||
* Entering user-defined LogP and pKa values allows the researcher to model the limiting effect of lipophilicity and ionization on intestinal permeation rate, thus providing a straightforward route for property-based design of oral drugs. | * Entering user-defined LogP and pKa values allows the researcher to model the limiting effect of lipophilicity and ionization on intestinal permeation rate, thus providing a straightforward route for property-based design of oral drugs. | ||
* Displays the experimental values of the relevant properties for up to 3 similar structures from the training set along with each Caco-2 permeability prediction.<br /> | |||
* Displays the experimental values of the relevant properties for up to 3 similar structures from | |||
==Interface== | ==Interface== | ||
Revision as of 07:37, 25 May 2012
Overview
Intestinal permeability of drugs together with solubility are the two key factors affecting their oral bioavailability. Absorption module provides accurate predictions of passive intestinal permeability (on jejunal and Caco-2 scales) and extent of oral absorption (%HIA) of drug candidates. These predictions based on intuitive, easily interpretable physicochemical models enable the researchers to rank and select lead compounds according to their permeability across intestinal barrier and to exclude candidates exhibiting extremely poor absorption at the earliest stages.
Caco-2 module contains a mechanistic predictive model of compound permeability across Caco-2 cell monolayer at different experimental conditions. The predictive algorithm was developed using data from >800 experiments with nearly 600 compounds. The resulting model is ionization-specific and also takes into account permeabilities via transcellular and paracellular routes. It uses essential physicochemical properties such as lipophilicity, ionization constants, number of H-Bond donors and molecular size (calculated or experimental if available) as inputs.
Features
- Calculates the extent of passive permeability of analyzed compounds across Caco-2 cell monolayers at specified pH and stirring conditions using physicochemical property values such as lipophilicity (LogP) and ionization (pKa) as inputs.
- Gives an estimate of relative contributions of different transport routes to Caco-2 permeability.
- Allows entering experimentally measured physicochemical properties instead of automatically calculated values to improve the quality of predictions. The experimental conditions (pH and stirring rate) may also be adjusted manually for full flexibility of the simulation.
- Entering user-defined LogP and pKa values allows the researcher to model the limiting effect of lipophilicity and ionization on intestinal permeation rate, thus providing a straightforward route for property-based design of oral drugs.
- Displays the experimental values of the relevant properties for up to 3 similar structures from the training set along with each Caco-2 permeability prediction.
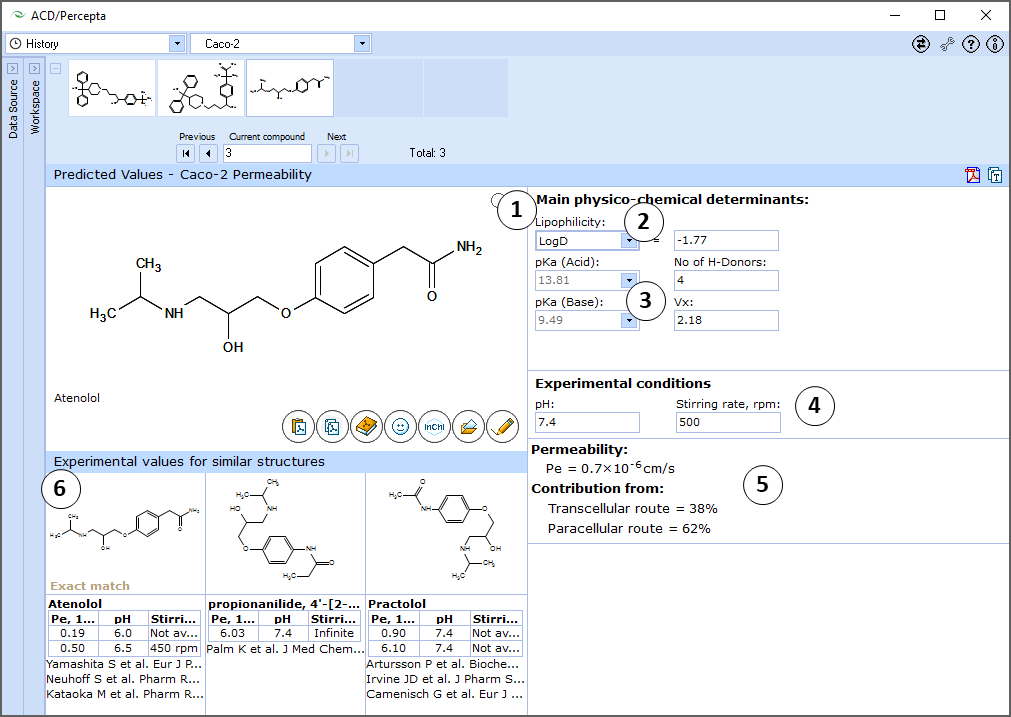
Interface
- Calculated logP, number of H-Bond donors and McGowan volume. Click and type a new value to model the limiting effect of lipophilicity, H-Bonding acidity and molecular size on Caco-2 permeability
- Calculated acid and base ionization constants. Click to select or type new pKa(acid) and pKa(base) values to model the limiting effect of ionization constants on Caco-2 permeability.
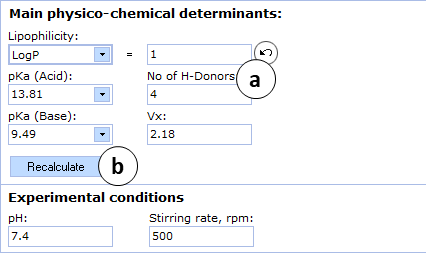
- a. Click to reverse to an automatically calculated property value (logP in this picture) for a compound and to recalculate the Caco-2 permeability
- b. Click to recalculate the Caco-2 permeability using the currently specified parameter values
- Experimental conditions used in simulation. Type in new values to change the pH or stirring rate (rpm)
- Estimated value of Caco-2 permeability (Pe, cm/s) and the relative contributions of transcellular and paracellular pathways, calculated as a function of compound structure and provided parameters
- Up to 3 most similar structures in the Absorption DB with experimental values and references
Technical information
Only a short summary of main technical aspects of Absorption predictor is given here. For a more detailed description of the modeling approach and underlying theory please refer to Reynolds, D. P., et al. J Pharm Sci. 2009; In press. [1]
Calculated quantitative parameters
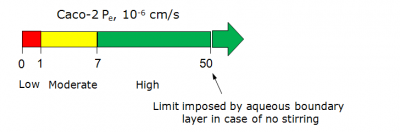
The main output of the Absorption module is the maximum achievable extent of human intestinal absorption (when solubility is not a limiting factor) expressed as a percentage value and denoted %HIA. Typically, compounds exhibiting %HIA > 70% are considered well absorbed, those with %HIA < 30% - poorly absorbed, while values in the range 30 - 70% represent moderate absorption. Intestinal permeability rates corresponding to the respective %HIA values are calibrated on several scales and given as:
- Effective jejunal permeability coefficients at pH 6.5 (Pe, 10-4 cm/s)
- Absorption rate constants (ka, min-1).
Additionally, Absorption\Caco-2 module presents effective permeability coefficients in Caco-2 monolayers (Pe, 10-6 cm/s) at user-defined conditions (pH and stirring rate).
Descriptors & Modeling Method
Descriptors used for modeling included key physicochemical properties - octanol/water LogP of neutral species as a determinant of lipophilicity, ion form fractions at pH 6.5 calculated from the respective pKa values, number of hydrogen bond donors in the molecule, and McGowan characetristic volume reflecting molecular size. All physicochemical parameter values were calculated with Algorithm Builder 1.8.
Due to the evidence of sigmoidal relationship between fraction absorbed (represented by experimental data) and actual permeation rates data were modeled using non-linear least squares regression. Fitting was performed in a multi-step approach, separate stages needed for:
- Determination of paracellular transport parameters
- Describing trancellular diffusion of non-electrolytes
- Estimating ionization dependence of intestinal absorption
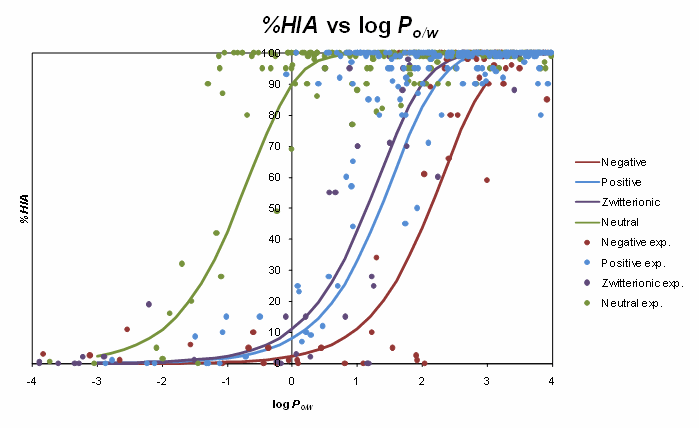
The sigmoid relationships between %HIA and octanol/water log P for various electrolyte classes are illustrated in the figure below. The sigmoids obtained for charged species are shifted relatively to the respective curve for non-electrolytes, although the shift is not as marked as could be expected if absorption was modeled by pH-dependent octanol/water distribution coefficient log D. This observation shows that using log D as a descriptor of in vivo membrane partitioning is not feasible. Ionization-specific analysis performed in this study is therefore necessary to devise appropriate corrections to log P values for correct prediction of passive membrane permeability.
Prediction accuracy
The final quantitative %HIA data set (cleaned of values distorted by P-gp efflux, facilitated diffusion and other side processes) that was used for modeling consisted of 567 %HIA values, mostly for marketed drugs or drug candidates. For validation purposes two independent test sets were compiled from recent publications on jejunal absorption of drugs. External validation sets represented another types of experimental data than that used in model development:
- The first set contained directly measured jejunal permeability coefficients (Peff) for 25 compounds extracted from Lennernäs, H. Xenobiotica. 2007;37(10-11):1015-51. [2]
- The second set was comprised of absorption rate constants (ka) for 22 molecules from the study by Linnankoski J. et al. J Med Chem. 2006;49(12):3674-81. [3].
The main advantage of such type of validation is the possibility to evaluate the intrinsic correctness of our model rather than just goodness of fit between experimental and predicted HIA.
Model performance on internal %HIA data set and external validation sets is summarized in the table below:
| Data set | N | R2 | RMSE |
|---|---|---|---|
| %HIA training set | 567 | 0.93 | 9.5% |
| log Peff validation set | 25 | 0.72 | 0.45 |
| log Ka validation set | 22 | 0.84 | 0.35 |
Note that RMSE for the training set is grayed since prediction error for percentage values (that are mostly concentrated on the ends of the scale) does not provide an unambigous measure of model quality, while the actual accuracy of predictions is best illustrated by model performance on external validation sets yielding RMSE about 0.4 log units which is close to the error of experimental determination.
Reference database
Absorption\Absorption DB module contains a browsable database of human intestinal absorption & bioavailability. The experimental data were compiled from reference pharmacokinetic tabulations and original articles, the main sources being "Therapeutic Drugs" (ed. by C. dollery), Goodman & Gilman's "The Pharmacological Basis of Therapeutics", and a compilation by Zhao, Y. H. et al. J Pharm Sci. 2001;90(6):749-84. A qualitative assignment of absorption category (good, moderate, poor) is provided for each compound in the database along with comments regarding the quantitative extent of absorption/bioavailability, presence of carrier-mediated transport, etc. where available.
Transport mechanisms
The predictive models comprising the Absorption module account for passive transport of analyzed compounds across intestinal barrier. In order to propose a clear physicochemical explanation of passive diffusion process, the data used for model development (both %HIA and Caco-2 permeability sets containing about 600 data points) were thoroughly evaluated to exclude values affected by enzymatic efflux or influx. For such compounds special alerts are displayed in the Bioavailability module indicating that expected oral bioavailability/absorption values for these molecules may be higher or lower due to presence of carrier-mediated processes.