Impurity Profiling: Difference between revisions
Updated for 2023 release |
|||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
Impurity profiling module is a result of the collaboration between ACD/Labs and FDA Center for Food Safety and Nutrition (CFSAN). Evaluation of genotoxic and/or carcinogenic potential is based on a battery of probabilistic models for bioassays reflecting different mechanisms of hazardous activity. A knowledge-based expert system identifies potentially hazardous structural fragments that could be responsible for carcinogenic activity of the test molecule. | Impurity profiling module is a result of the collaboration between ACD/Labs and FDA Center for Food Safety and Nutrition (CFSAN). Evaluation of genotoxic and/or carcinogenic potential is based on a battery of probabilistic models for bioassays reflecting different mechanisms of hazardous activity. A knowledge-based expert system identifies potentially hazardous structural fragments that could be responsible for carcinogenic activity of the test molecule. | ||
The toxicity predictions in the | The toxicity predictions in the impurity profiling package offer greater insight into the safety of impurities, providing detailed information on toxic endpoints, reflecting various mechanisms of hazardous activity including: | ||
* Mutagenicity (Ames test, Mouse Lymphoma Assay, and other standard assays) | * Mutagenicity (Ames test, Mouse Lymphoma Assay, and other standard assays) | ||
* Clastogenicity (Micronucleus test, Chromosomal Aberrations) | * Clastogenicity (Micronucleus test, Chromosomal Aberrations) | ||
| Line 11: | Line 11: | ||
* Endocrine disruption mechanisms (estrogen receptor binding) | * Endocrine disruption mechanisms (estrogen receptor binding) | ||
The impurities package offers probabilistic predictive models for 21 different endpoints that cover various mechanisms of hazardous activity presented above. These predictors are supplemented with a knowledge-based expert system that identifies potentially hazardous structural fragments that could be responsible for genotoxic and/or carcinogenic activity of the compound of interest. | The impurities package offers probabilistic predictive models for 21 different endpoints that cover various mechanisms of hazardous activity presented above. These predictors are supplemented with a knowledge-based expert system that identifies potentially hazardous structural fragments that could be responsible for genotoxic and/or carcinogenic activity of the compound of interest. | ||
The set of property predictors is supplemented with an automatic classification system that classifies impurities by their genotoxic and carcinogenic potential according to ICH M7 Guidelines by European Medicines Agency. This classifier can aid the users with interpretation of the prediction results and preparation of compound safety reports for submission to regulatory authorities. | |||
<br /> | <br /> | ||
| Line 18: | Line 21: | ||
* Gain insight into the possible mechanisms of toxic effects | * Gain insight into the possible mechanisms of toxic effects | ||
* See a display of up to 5 similar structures with experimental results in relevant bioassays | * See a display of up to 5 similar structures with experimental results in relevant bioassays | ||
* Generate PDF reports in a variety of formats including ICH M7 Classification report providing full details regarding the assignment of a particular class and recommendations regarding appropriate control measures for that class of impurities. | |||
<br /> | <br /> | ||
| Line 23: | Line 27: | ||
<br /> | <br /> | ||
[[Image: | [[Image:Genotoxicity_impurity_profiling.png|center]] | ||
<br /> | <br /> | ||
# View the ICH M7 Class assigned to the compound of interest. Hover over the "i" icon to display a tooltip with a listing of evidence contributing to the classification. | |||
# Hover over the name of an alert to highlight the alerting group on the structure of the molecule | # Hover over the name of an alert to highlight the alerting group on the structure of the molecule | ||
# The list of all alerting groups found in the molecule. Each alert is supplied with statistical data regarding distrubution of positive and negative compounds possessing this hazardous fragment in all considered databases, along with the respective z-scores. Z-scores show whether the presence of the fragment leads to a statistically significant increase in proportion of compounds with a positive test result for a particular assay. This information provides further evidence regarding the possible mechanisms of action. | # The list of all alerting groups found in the molecule. Each alert is supplied with statistical data regarding distrubution of positive and negative compounds possessing this hazardous fragment in all considered databases, along with the respective z-scores. Z-scores show whether the presence of the fragment leads to a statistically significant increase in proportion of compounds with a positive test result for a particular assay. This information provides further evidence regarding the possible mechanisms of action. | ||
| Line 35: | Line 40: | ||
<div class="mw-collapsible | <div class="mw-collapsible"> | ||
==Technical information== | ==Technical information== | ||
| Line 41: | Line 46: | ||
<div class="mw-collapsible-content"> | <div class="mw-collapsible-content"> | ||
ACD/Labs Package for Toxicity Screening of Impurities provides a battery of in silico tests to accurately assess the genotoxic and carcinogenic potential of impurities and degradants, found to be below the threshold of toxicological concern in drug products, helping companies remain compliant with regulatory submission requirements. | ACD/Labs Package for Toxicity Screening of Impurities provides a battery of ''in silico'' tests to accurately assess the genotoxic and carcinogenic potential of impurities and degradants, found to be below the threshold of toxicological concern in drug products, helping companies remain compliant with regulatory submission requirements.<br> | ||
Profile impurities using predictions for genotoxic and carcinogenic endpoints, quickly determine if an impurity is likely to pose a safety risk, and identify potentially hazardous structural fragments responsible for toxic activity. | Profile impurities using predictions for genotoxic and carcinogenic endpoints, quickly determine if an impurity is likely to pose a safety risk, and identify potentially hazardous structural fragments responsible for toxic activity. | ||
<br /> | |||
The expert system contains a list of 67 alerting groups of toxicophores, 53 of which account for point mutational and/or clastogenic mechanisms of DNA damage, while the remaining 14 substructures detect carcinogens acting by non-genotoxic mechanisms. The expert system was able to recognize >94% of mutagens in ACD/Ames test database, and >90% of compounds marked as potent carcinogens in the FDA's OFAS Food-Additive Knowledgebase. | The expert system contains a list of 67 alerting groups of toxicophores, 53 of which account for point mutational and/or clastogenic mechanisms of DNA damage, while the remaining 14 substructures detect carcinogens acting by non-genotoxic mechanisms. The expert system was able to recognize >94% of mutagens in ACD/Ames test database, and >90% of compounds marked as potent carcinogens in the FDA's OFAS Food-Additive Knowledgebase. | ||
<br /> | |||
===ICH M7 Classification=== | |||
The impurities classification algorithm has been devised in accordance with "ICH guideline M7(R1) on assessment and control of DNA reactive (mutagenic) impurities in pharmaceuticals to limit potential carcinogenic risk" by European Medicines Agency [https://www.ema.europa.eu/en/documents/scientific-guideline/ich-guideline-m7r1-assessment-control-dna-reactive-mutagenic-impurities-pharmaceuticals-limit_en.pdf]. Specifically, this document considers 5 classes of impurities: | |||
{| border="1" class="wikitable" | |||
|- | |||
! Class !! Brief definition | |||
|- | |||
| style="text-align: center" | 1 || Known mutagenic carcinogens | |||
|- | |||
| style="text-align: center" | 2 || Known mutagens with unknown carcinogenic potential | |||
|- | |||
| style="text-align: center" | 3 || Alerting structure, unrelated to the structure of the drug substance | |||
|- | |||
| style="text-align: center" | 4 || Alerting structure, same alert in drug substance or compounds related to the drug substance | |||
|- | |||
| style="text-align: center" | 5 || No structural alerts, or alerting structure with sufficient data to demonstrate lack of mutagenicity or carcinogenicity | |||
|} | |||
The classification algorithm has been developed with the intent to mimic the logic of human expert evaluation. Each classification output is supplemented with reasoning that had led to assignment of a particular class and recommendations regarding appropriate control measures for that class of impurities. When classification cannot be made on the basis of available experimental data alone, further evaluation is performed using WOE (weight of evidence) approach involving: | |||
* The probability of hazardous effects reported by statistical models and confidence of predictions | |||
* Presence of alerting groups known from the literature | |||
* Evidence from experimental data for the most similar compounds from the built-in database | |||
* Other mitigating factors | |||
'''''Note:''''' Currently, Percepta can only assign Class 4 when ICH M7 Classification is calculated in Spreadsheet workspace with indicated *ID of the parent compound in the active project. Calculations in the Expert module UI (and in Spreadsheet with provided *ID = 0) consider the compounds one by one without account for potential parent-derivative relationships. Also, when a definitive classification cannot be made, ICH M7 Class is reported as Inconclusive (rendered as 0 in Spreadsheet workspace) – this should be treated similarly to Class 3 compounds, as requiring further attention. | |||
=== Experimental Data === | |||
A complete list of modeled endpoints is provided in the Table, while the data sources are briefly described below.<br /> | |||
'''Genetic toxicity''': data sets for standard assays reflecting different mechanisms of genetic damage were obtained from the FDA. Gene mutation tests and techniques detecting clastogenic/aneugenic effects are included. Data was collected from EPA GENE-TOX database [https://www.nlm.nih.gov/databases/download/genetox.html] and scientific literature. | |||
'''Carcinogenicity''': results of chronic (two-year term) carcinogenicity studies in rats and mice were received from FDA. This data was based on NTP technical reports, IARC monographs, Carcinogenic Potency DataBase [https://pubmed.ncbi.nlm.nih.gov/15800034] and other publicly available sources. Raw data was converted to binary classification using a weight of evidence (WOE) approach [https://pubmed.ncbi.nlm.nih.gov/17207562]. Classification using the WOE threshold corresponding to “potent carcinogens” was used to build the models in the current study. | |||
'''Reproductive toxicity''': experimental data characterizing the potential for endocrine system disruption due to Estrogen receptor α binding were acquired from ChEMBL database [https://www.ebi.ac.uk/chembl] (Target ID 206). Compounds were classified as binders/non-binders on the basis of their relative binding affinities (RBA) compared to reference ligand estradiol. Two cut-offs were used: LogRBA > -3 (“general binding”), and LogRBA > 0 (“strong binding”)<br /> | |||
<br /> | |||
A | {| border="1" class="wikitable" style="margin: 1em auto 1em auto; text-align: left" | ||
|+ Table. A list of bioassays and dataset sizes included in ACD/Impurity Profiler. | |||
|- | |||
! width="100" | Mechanism | |||
! width="150" | Test system | |||
! width="150" | Endpoint | |||
! width="100" style="text-align: center;" | N (Overall) | |||
! width="100" style="text-align: center;" | N (Positives) | |||
! width="100" style="text-align: center;" | % Positives | |||
|- | |||
| style="background: #EEF4E4" rowspan="8" | Mutagenicity | |||
| style="background: #EEF4E4" rowspan="3" | Prokaryote | |||
| style="background: #EEF4E4" | Composite | |||
| style="background: #EEF4E4; text-align: center;" | 7953 | |||
| style="background: #EEF4E4; text-align: center;" | 4003 | |||
| style="background: #EEF4E4; text-align: center;" | 50.3% | |||
|- | |||
| style="background: #EEF4E4" | Salmonella | |||
| style="background: #EEF4E4; text-align: center;" | 7826 | |||
| style="background: #EEF4E4; text-align: center;" | 3875 | |||
| style="background: #EEF4E4; text-align: center;" | 49.5% | |||
|- | |||
| style="background: #EEF4E4" | Escherichia | |||
| style="background: #EEF4E4; text-align: center;" | 1479 | |||
| style="background: #EEF4E4; text-align: center;" | 386 | |||
| style="background: #EEF4E4; text-align: center;" | 26.1% | |||
|- | |||
| style="background: #EEF4E4" rowspan="5" | Eukaryote | |||
| style="background: #EEF4E4" | Composite | |||
| style="background: #EEF4E4; text-align: center;" | 2901 | |||
| style="background: #EEF4E4; text-align: center;" | 1592 | |||
| style="background: #EEF4E4; text-align: center;" | 54.9% | |||
|- | |||
| style="background: #EEF4E4" | Yeast | |||
| style="background: #EEF4E4; text-align: center;" | 658 | |||
| style="background: #EEF4E4; text-align: center;" | 347 | |||
| style="background: #EEF4E4; text-align: center;" | 52.7% | |||
|- | |||
| style="background: #EEF4E4" | Drosophila | |||
| style="background: #EEF4E4; text-align: center;" | 600 | |||
| style="background: #EEF4E4; text-align: center;" | 293 | |||
| style="background: #EEF4E4; text-align: center;" | 48.8% | |||
|- | |||
| style="background: #EEF4E4" | Mouse Lymphoma Assay | |||
| style="background: #EEF4E4; text-align: center;" | 1272 | |||
| style="background: #EEF4E4; text-align: center;" | 763 | |||
| style="background: #EEF4E4; text-align: center;" | 60.0% | |||
|- | |||
| style="background: #EEF4E4" | CHO/CHL all loci | |||
| style="background: #EEF4E4; text-align: center;" | 1229 | |||
| style="background: #EEF4E4; text-align: center;" | 585 | |||
| style="background: #EEF4E4; text-align: center;" | 47.6% | |||
|- | |||
| style="background: #FCEEE3" rowspan="3" | Clastogenicity | |||
| style="background: #FCEEE3" rowspan="2" | Chromosome aberrations | |||
| style="background: #FCEEE3" | ''In vitro'' | |||
| style="background: #FCEEE3; text-align: center;" | 2034 | |||
| style="background: #FCEEE3; text-align: center;" | 941 | |||
| style="background: #FCEEE3; text-align: center;" | 46.3% | |||
|- | |||
| style="background: #FCEEE3" | ''In vivo'' | |||
| style="background: #FCEEE3; text-align: center;" | 441 | |||
| style="background: #FCEEE3; text-align: center;" | 133 | |||
| style="background: #FCEEE3; text-align: center;" | 30.2% | |||
|- | |||
| style="background: #FCEEE3" | Micronucleus test in rodents | |||
| style="background: #FCEEE3" | ''In vivo'' | |||
| style="background: #FCEEE3; text-align: center;" | 1299 | |||
| style="background: #FCEEE3; text-align: center;" | 403 | |||
| style="background: #FCEEE3; text-align: center;" | 31.0% | |||
|- | |||
| style="background: #F4E1E1" | DNA damage | |||
| style="background: #F4E1E1" | Unscheduled DNA synthesis | |||
| style="background: #F4E1E1" | ''In vivo/in vitro'' | |||
| style="background: #F4E1E1; text-align: center;" | 593 | |||
| style="background: #F4E1E1; text-align: center;" | 166 | |||
| style="background: #F4E1E1; text-align: center;" | 28.0% | |||
|- | |||
| style="background: #ECE7EF" rowspan="5" | Carcinogenicity | |||
| style="background: #ECE7EF" | Rodent | |||
| style="background: #ECE7EF" | Composite | |||
| style="background: #ECE7EF; text-align: center;" | 2211 | |||
| style="background: #ECE7EF; text-align: center;" | 674 | |||
| style="background: #ECE7EF; text-align: center;" | 30.5% | |||
|- | |||
| style="background: #ECE7EF" rowspan="2" | Rat | |||
| style="background: #ECE7EF" | Male | |||
| style="background: #ECE7EF; text-align: center;" | 1818 | |||
| style="background: #ECE7EF; text-align: center;" | 647 | |||
| style="background: #ECE7EF; text-align: center;" | 35.6% | |||
|- | |||
| style="background: #ECE7EF" | Female | |||
| style="background: #ECE7EF; text-align: center;" | 1793 | |||
| style="background: #ECE7EF; text-align: center;" | 635 | |||
| style="background: #ECE7EF; text-align: center;" | 35.4% | |||
|- | |||
| style="background: #ECE7EF" rowspan="2" | Mouse | |||
| style="background: #ECE7EF" | Male | |||
| style="background: #ECE7EF; text-align: center;" | 1669 | |||
| style="background: #ECE7EF; text-align: center;" | 556 | |||
| style="background: #ECE7EF; text-align: center;" | 33.3% | |||
|- | |||
| style="background: #ECE7EF" | Female | |||
| style="background: #ECE7EF; text-align: center;" | 1727 | |||
| style="background: #ECE7EF; text-align: center;" | 561 | |||
| style="background: #ECE7EF; text-align: center;" | 32.5% | |||
|- | |||
| style="background: #E2EAF4" rowspan="2" | Reproductive toxicity | |||
| style="background: #E2EAF4" rowspan="2" | Estrogen receptor binding | |||
| style="background: #E2EAF4" | LogRBA > 0 | |||
| style="background: #E2EAF4; text-align: center;" | 3423 | |||
| style="background: #E2EAF4; text-align: center;" | 1488 | |||
| style="background: #E2EAF4; text-align: center;" | 43.5% | |||
|- | |||
| style="background: #E2EAF4" | LogRBA > -3 | |||
| style="background: #E2EAF4; text-align: center;" | 3423 | |||
| style="background: #E2EAF4; text-align: center;" | 2549 | |||
| style="background: #E2EAF4; text-align: center;" | 74.5% | |||
|} | |||
<br /> | |||
=== Methods === | |||
Probabilistic predictive models for all considered endpoints were developed using GALAS modeling methodology [https://pubmed.ncbi.nlm.nih.gov/20814717]. Each GALAS model consists of two parts: | |||
# Global (baseline) model that reflects general trends in the property of interest. Baseline models were built using binomial PLS method based on fragmental descriptors. | |||
# Local corrections were applied to baseline predictions using a special similarity-based routine, after performing an analysis for the most similar compounds used in the training set. The local part of the model provides the basis for the calculation of the Reliability index (RI), a value ranging from 0 to 1 that provides a quantitative estimate of prediction accuracy. | |||
A single baseline model was derived for each group of endpoints representing the same mechanism of hazardous action. Such model reflects a “cumulative” toxicity potential of chemicals in these assays. Experimental values specific for a particular assay were used during the local part of the modeling to yield final GALAS model for that endpoint. <br /> | |||
=== Genotoxicity/Carcinogenicity Hazards === | |||
The | The knowledge-based expert system that identifies structural fragments potentially responsible for genotoxic effect is an extension of the previously described Ames mutagenicity hazards system [https://www.sciencedirect.com/science/article/abs/pii/S0378427408007054]. The list of alerting groups was augmented with structural moieties that are frequently present in compounds tested positive in chromosomal damage assays, eucaryote gene mutation tests, as well as in carcinogens acting by non-genotoxic (epigenetic) mechanisms. The final list included 67 structural alerts, 14 of which represent epigenetic carcinogens (androgens, peroxisome proliferators, etc.) [https://www.sciencedirect.com/science/article/abs/pii/S0378427411005492].<br /> | ||
Overall, the expert system was able to detect 94% of mutagens in the Ames test DB and 90% of compounds labeled as potent carcinogens by FDA.<br /> | |||
The alert list is not limited to directly acting substructures, such as planar polycyclic arenes, aromatic amines, quinones, N-nitro and N-nitroso groups, but also includes various fragments that may undergo biotransformation to reactive intermediates. As an example, troglizatone, a thiazolidinedione class antidiabetic drug, was classified by the FDA as a potent carcinogen and has since been withdrawn from the USA market. The carcinogenic effect of this drug is mediated by several reactive metabolites. In human liver microsomes, the chromane ring of troglitazone is metabolized by CYP3A4 to form quinone and quinone-methide products. Furthermore, oxidative cleavage of thiazolidinedione ring results in a reactive sulfenic acid metabolite that also contains an isocyanate moiety [https://pubmed.ncbi.nlm.nih.gov/20869346].<br /> | |||
<br /> | |||
[1] Matthews EJ et al. Regul Toxicol Pharmacol. 2007, 47, 115. | <!--=== References === | ||
[2] Gold LS et al. Toxicol Sci. 2005, 85, 747. | [1] Matthews EJ et al. Regul Toxicol Pharmacol. 2007, 47, 115.<br> | ||
[3] ChEMBL DB (https://www.ebi.ac.uk/chembldb). Target ID 206. | [2] Gold LS et al. Toxicol Sci. 2005, 85, 747.<br> | ||
[4] Didziapetris R et al. J Comput Aided Mol Des. 2010, 24, 891. | [3] ChEMBL DB (https://www.ebi.ac.uk/chembldb). Target ID 206.<br> | ||
[5] Didziapetris R et al. Toxicol Lett. 2008, 180, S152. | [4] Didziapetris R et al. J Comput Aided Mol Des. 2010, 24, 891.<br> | ||
[6] Mansuy D & Dansette PM. Arch Biochem Biophys. 2011, 507, 1745. | [5] Didziapetris R et al. Toxicol Lett. 2008, 180, S152.<br> | ||
[6] Mansuy D & Dansette PM. Arch Biochem Biophys. 2011, 507, 1745.<br> | |||
--> | |||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 10:53, 26 July 2023
Overview
Impurity profiling module is a result of the collaboration between ACD/Labs and FDA Center for Food Safety and Nutrition (CFSAN). Evaluation of genotoxic and/or carcinogenic potential is based on a battery of probabilistic models for bioassays reflecting different mechanisms of hazardous activity. A knowledge-based expert system identifies potentially hazardous structural fragments that could be responsible for carcinogenic activity of the test molecule.
The toxicity predictions in the impurity profiling package offer greater insight into the safety of impurities, providing detailed information on toxic endpoints, reflecting various mechanisms of hazardous activity including:
- Mutagenicity (Ames test, Mouse Lymphoma Assay, and other standard assays)
- Clastogenicity (Micronucleus test, Chromosomal Aberrations)
- DNA damage (Unscheduled DNA Synthesis)
- Carcinogenicity (FDA rodent carcinogenicity data)
- Endocrine disruption mechanisms (estrogen receptor binding)
The impurities package offers probabilistic predictive models for 21 different endpoints that cover various mechanisms of hazardous activity presented above. These predictors are supplemented with a knowledge-based expert system that identifies potentially hazardous structural fragments that could be responsible for genotoxic and/or carcinogenic activity of the compound of interest.
The set of property predictors is supplemented with an automatic classification system that classifies impurities by their genotoxic and carcinogenic potential according to ICH M7 Guidelines by European Medicines Agency. This classifier can aid the users with interpretation of the prediction results and preparation of compound safety reports for submission to regulatory authorities.
Features
- Predict the genotoxic and carcinogenic effects of an impurity from simple structure input (name, 2D structure, SMILES string), with a reliability index generated by the probabilistic models
- Identify potentially hazardous structural fragments responsible for carcinogenic and genotoxic activity
- Gain insight into the possible mechanisms of toxic effects
- See a display of up to 5 similar structures with experimental results in relevant bioassays
- Generate PDF reports in a variety of formats including ICH M7 Classification report providing full details regarding the assignment of a particular class and recommendations regarding appropriate control measures for that class of impurities.
Interface
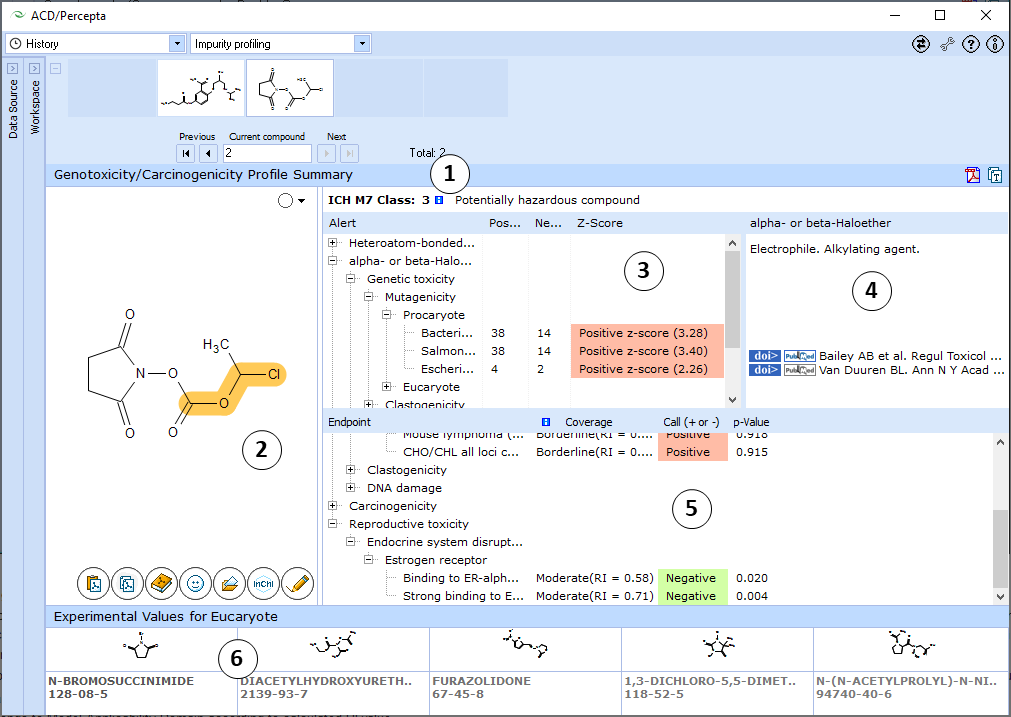
- View the ICH M7 Class assigned to the compound of interest. Hover over the "i" icon to display a tooltip with a listing of evidence contributing to the classification.
- Hover over the name of an alert to highlight the alerting group on the structure of the molecule
- The list of all alerting groups found in the molecule. Each alert is supplied with statistical data regarding distrubution of positive and negative compounds possessing this hazardous fragment in all considered databases, along with the respective z-scores. Z-scores show whether the presence of the fragment leads to a statistically significant increase in proportion of compounds with a positive test result for a particular assay. This information provides further evidence regarding the possible mechanisms of action.
- Each hazardous fragment is provided with a short description of its mechanism of action and literature references.
- The output of probabilistic models is presented in the form of a tree view, where the nodes corresponding to individual endpoints are grouped into higher level nodes according to species/test system and mechanism of action. The output for each endpoint consists of the following parts:
- p-value – probability that a compound will result in a positive test in the respective assay
- Coverage – an indication whether the compound belongs to Model Applicability Domain according to calculated RI value
- Call – (+ or –) if the compound can be reliably classified on the basis of p and RI values, “Undefined” otherwise.
- Clicking on a tree node brings up 5 most similar structures in the respective training set with names, CAS numbers and experimental results (positive or negative, as well as quantitative TD50 values and tumour target sites in case of carcinogenicity)
Technical information
ACD/Labs Package for Toxicity Screening of Impurities provides a battery of in silico tests to accurately assess the genotoxic and carcinogenic potential of impurities and degradants, found to be below the threshold of toxicological concern in drug products, helping companies remain compliant with regulatory submission requirements.
Profile impurities using predictions for genotoxic and carcinogenic endpoints, quickly determine if an impurity is likely to pose a safety risk, and identify potentially hazardous structural fragments responsible for toxic activity.
The expert system contains a list of 67 alerting groups of toxicophores, 53 of which account for point mutational and/or clastogenic mechanisms of DNA damage, while the remaining 14 substructures detect carcinogens acting by non-genotoxic mechanisms. The expert system was able to recognize >94% of mutagens in ACD/Ames test database, and >90% of compounds marked as potent carcinogens in the FDA's OFAS Food-Additive Knowledgebase.
ICH M7 Classification
The impurities classification algorithm has been devised in accordance with "ICH guideline M7(R1) on assessment and control of DNA reactive (mutagenic) impurities in pharmaceuticals to limit potential carcinogenic risk" by European Medicines Agency [1]. Specifically, this document considers 5 classes of impurities:
| Class | Brief definition |
|---|---|
| 1 | Known mutagenic carcinogens |
| 2 | Known mutagens with unknown carcinogenic potential |
| 3 | Alerting structure, unrelated to the structure of the drug substance |
| 4 | Alerting structure, same alert in drug substance or compounds related to the drug substance |
| 5 | No structural alerts, or alerting structure with sufficient data to demonstrate lack of mutagenicity or carcinogenicity |
The classification algorithm has been developed with the intent to mimic the logic of human expert evaluation. Each classification output is supplemented with reasoning that had led to assignment of a particular class and recommendations regarding appropriate control measures for that class of impurities. When classification cannot be made on the basis of available experimental data alone, further evaluation is performed using WOE (weight of evidence) approach involving:
- The probability of hazardous effects reported by statistical models and confidence of predictions
- Presence of alerting groups known from the literature
- Evidence from experimental data for the most similar compounds from the built-in database
- Other mitigating factors
Note: Currently, Percepta can only assign Class 4 when ICH M7 Classification is calculated in Spreadsheet workspace with indicated *ID of the parent compound in the active project. Calculations in the Expert module UI (and in Spreadsheet with provided *ID = 0) consider the compounds one by one without account for potential parent-derivative relationships. Also, when a definitive classification cannot be made, ICH M7 Class is reported as Inconclusive (rendered as 0 in Spreadsheet workspace) – this should be treated similarly to Class 3 compounds, as requiring further attention.
Experimental Data
A complete list of modeled endpoints is provided in the Table, while the data sources are briefly described below.
Genetic toxicity: data sets for standard assays reflecting different mechanisms of genetic damage were obtained from the FDA. Gene mutation tests and techniques detecting clastogenic/aneugenic effects are included. Data was collected from EPA GENE-TOX database [2] and scientific literature.
Carcinogenicity: results of chronic (two-year term) carcinogenicity studies in rats and mice were received from FDA. This data was based on NTP technical reports, IARC monographs, Carcinogenic Potency DataBase [3] and other publicly available sources. Raw data was converted to binary classification using a weight of evidence (WOE) approach [4]. Classification using the WOE threshold corresponding to “potent carcinogens” was used to build the models in the current study.
Reproductive toxicity: experimental data characterizing the potential for endocrine system disruption due to Estrogen receptor α binding were acquired from ChEMBL database [5] (Target ID 206). Compounds were classified as binders/non-binders on the basis of their relative binding affinities (RBA) compared to reference ligand estradiol. Two cut-offs were used: LogRBA > -3 (“general binding”), and LogRBA > 0 (“strong binding”)
| Mechanism | Test system | Endpoint | N (Overall) | N (Positives) | % Positives |
|---|---|---|---|---|---|
| Mutagenicity | Prokaryote | Composite | 7953 | 4003 | 50.3% |
| Salmonella | 7826 | 3875 | 49.5% | ||
| Escherichia | 1479 | 386 | 26.1% | ||
| Eukaryote | Composite | 2901 | 1592 | 54.9% | |
| Yeast | 658 | 347 | 52.7% | ||
| Drosophila | 600 | 293 | 48.8% | ||
| Mouse Lymphoma Assay | 1272 | 763 | 60.0% | ||
| CHO/CHL all loci | 1229 | 585 | 47.6% | ||
| Clastogenicity | Chromosome aberrations | In vitro | 2034 | 941 | 46.3% |
| In vivo | 441 | 133 | 30.2% | ||
| Micronucleus test in rodents | In vivo | 1299 | 403 | 31.0% | |
| DNA damage | Unscheduled DNA synthesis | In vivo/in vitro | 593 | 166 | 28.0% |
| Carcinogenicity | Rodent | Composite | 2211 | 674 | 30.5% |
| Rat | Male | 1818 | 647 | 35.6% | |
| Female | 1793 | 635 | 35.4% | ||
| Mouse | Male | 1669 | 556 | 33.3% | |
| Female | 1727 | 561 | 32.5% | ||
| Reproductive toxicity | Estrogen receptor binding | LogRBA > 0 | 3423 | 1488 | 43.5% |
| LogRBA > -3 | 3423 | 2549 | 74.5% |
Methods
Probabilistic predictive models for all considered endpoints were developed using GALAS modeling methodology [6]. Each GALAS model consists of two parts:
- Global (baseline) model that reflects general trends in the property of interest. Baseline models were built using binomial PLS method based on fragmental descriptors.
- Local corrections were applied to baseline predictions using a special similarity-based routine, after performing an analysis for the most similar compounds used in the training set. The local part of the model provides the basis for the calculation of the Reliability index (RI), a value ranging from 0 to 1 that provides a quantitative estimate of prediction accuracy.
A single baseline model was derived for each group of endpoints representing the same mechanism of hazardous action. Such model reflects a “cumulative” toxicity potential of chemicals in these assays. Experimental values specific for a particular assay were used during the local part of the modeling to yield final GALAS model for that endpoint.
Genotoxicity/Carcinogenicity Hazards
The knowledge-based expert system that identifies structural fragments potentially responsible for genotoxic effect is an extension of the previously described Ames mutagenicity hazards system [7]. The list of alerting groups was augmented with structural moieties that are frequently present in compounds tested positive in chromosomal damage assays, eucaryote gene mutation tests, as well as in carcinogens acting by non-genotoxic (epigenetic) mechanisms. The final list included 67 structural alerts, 14 of which represent epigenetic carcinogens (androgens, peroxisome proliferators, etc.) [8].
Overall, the expert system was able to detect 94% of mutagens in the Ames test DB and 90% of compounds labeled as potent carcinogens by FDA.
The alert list is not limited to directly acting substructures, such as planar polycyclic arenes, aromatic amines, quinones, N-nitro and N-nitroso groups, but also includes various fragments that may undergo biotransformation to reactive intermediates. As an example, troglizatone, a thiazolidinedione class antidiabetic drug, was classified by the FDA as a potent carcinogen and has since been withdrawn from the USA market. The carcinogenic effect of this drug is mediated by several reactive metabolites. In human liver microsomes, the chromane ring of troglitazone is metabolized by CYP3A4 to form quinone and quinone-methide products. Furthermore, oxidative cleavage of thiazolidinedione ring results in a reactive sulfenic acid metabolite that also contains an isocyanate moiety [9].