Oral Bioavailability: Difference between revisions
No edit summary |
|||
| (19 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
==Overview== | ==Overview== | ||
Oral Bioavailability module predicts the fraction of the specified drug dose that reaches systemic circulation after oral administration (''%F''). For calculation of quantitative ''%F'' values and exploring the dose-dependence of bioavailability, Oral bioavailability module uses the same kind of absorption simulation that is implemented in [[PK_Explorer|ACD/PK Explorer]]. | |||
<br/> | |||
<br/> | |||
===Features=== | ===Features=== | ||
* Predicts ''%F'' after oral administration with the possibility to explore dose-dependence of bioavailability | |||
<br /> | * Predicts a number of endpoints that affect oral bioavailability: | ||
** Solubility (dose/solubility ratio) | |||
** Stability in acidic media | |||
** Intestinal membrane permeability by passive or active transport | |||
** Likelihood of P-gp efflux | |||
** First pass metabolism in the liver | |||
* Visualizes the contributions of underlying properties with traffic-lights (green = good, red = problematic) for easy interpretation | |||
* Displays experimental ''%F'' values for up to 5 similar structures from Bioavailability DB along with literature references. | |||
<br/> | |||
==Interface== | |||
[[Image:Oral_bioavailability.png|center]] | |||
<br> | |||
<ol> | <ol> | ||
<li>50 mg is the default oral drug dose used in calculations. Enter any desired value to explore the effect of dose on oral bioavailability.<br> [[Image:Oral_bioavailability_simulation.png|right]] | |||
:a. Click the "Undo" button to reset the specified drug dose and to recalculate ''%F'' using the default settings. | |||
:b. Click to recalculate ''%F'' using the currently specified dose.</li><br> | |||
<li>Predicted ''%F'' (Oral) value.<br/>[[Image:Oral_Bioavailability_Ranges.png]]</li><br> | |||
<li>Click to see more details regarding the calculation of the particular property.</li><br> | |||
<li>Factors affecting oral bioavailability (see below for details).</li><br> | |||
<br /> | <li>Hover over a title to view a screentip with a short description.</li><br> | ||
<li>Up to 5 similar structures in the Bioavailability DB with experimental values and references.</li> | |||
</ol> | |||
<br/> | |||
====Traffic-lights system explanation==== | ====Traffic-lights system explanation==== | ||
:''<b>Solubility in gastrointestinal tract:</b>'' | |||
:* <b><font color="limegreen">Green</font></b> – good – dose/solubility ratio < 1. | |||
:* <b><font color="orange">Yellow</font></b> – moderate – dose/solubility ratio between 1 and 10. | |||
:* <b><font color="red">Red</font></b> – poor – dose/solubility ratio > 10. | |||
:''<b>Stability</b> – susceptibility to acid hydrolysis in stomach:'' | |||
:* <b><font color="red">Red</font></b> – only assigned to highly reactive compounds that decompose in stomach very quickly. Red light means that ''%F'' (oral) <10%, overriding all other predictions (''i.e.'', do not pay attention to predicted values in this particular case). | |||
:''<b>Passive absorption</b> – ability to cross human intestinal membrane by passive diffusion:'' | |||
:* <b><font color="red">Red</font></b> – intestinal passive absorption <30%. Poor bioavailability, as ''%F'' (oral) cannot exceed the extent of passive absorption. | |||
:* <b><font color="limegreen">Green</font></b> – good (>70%) passive absorption across intestinal barrier. Passive absorption does not affect ''%F''. | |||
:''<b>First-pass metabolism</b> – susceptibility to metabolic transformations catalyzed by enzymes in liver and intestine:'' | |||
:*<b><font color="red">Red</font></b> – high probability that first-pass metabolism is >50%. In this case ''%F'' (oral) is likely to be dose dependent and not to exceed 40%. | |||
:*<b><font color="limegreen">Green</font></b> – compound probably does not undergo significant first-pass metabolism. | |||
:''<b>P-gp efflux</b> – susceptibility to backward transport through intestinal membrane:'' | |||
:*<b><font color="red">Red</font></b> – compound is P-glycoprotein substrate. This effect is mostly important when compound is metabolized by CYP3A4 | |||
:''<b>Active transport</b> – susceptibility to active transport through intestinal membrane:'' | |||
:*<b><font color="limegreen">Green</font></b> – compound is actively transported by PepT1, ASBT or other enzymes. | |||
:*<b><font color="red">Red</font></b> light never appears, as this factor can only increase ''%F'' (oral). | |||
<br/> | |||
<br /> | |||
<div class="mw-collapsible"> | |||
<div class="mw-collapsible | |||
==Technical information== | ==Technical information== | ||
<br /> | <br/> | ||
<div class="mw-collapsible-content"> | <div class="mw-collapsible-content"> | ||
''' | ===Bioavailability DB=== | ||
'''Number of compounds:''' 788<br> | |||
'''Main sources of experimental data:''' | '''Main sources of experimental data:''' | ||
* Reference books: | * Reference books: | ||
** <i>Therapeutic Drugs</i>, Dolery, C., Ed. 2nd Edition, Churchill Livingstone, New York, NY, 1999 | ** <i>Therapeutic Drugs</i>, Dolery, C., Ed. 2nd Edition, Churchill Livingstone, New York, NY, 1999 | ||
** <i>Clarke's Isolation and Identification of Drugs</i>, Moffat, A.C., Jackson, J.V., Moss, M.S., Widdop, B., Eds. 2nd Edition, The Pharmaceutical Press, London, 1986 | ** <i>Clarke's Isolation and Identification of Drugs</i>, Moffat, A.C., Jackson, J.V., Moss, M.S., Widdop, B., Eds. 2nd Edition, The Pharmaceutical Press, London, 1986 | ||
* Various articles from peer-reviewed scientific journals | * Various articles from peer-reviewed scientific journals, including both detailed pharmacokinetic characterization studies (''i.e.'', usually only several compounds per article), and larger data compilations | ||
<br> | |||
===Simulation model=== | |||
The mathematical model that is used for simulations performed by PK Explorer and Oral Bioavailability modules is based on differential equations that consider solubility in the gastrointestinal tract, passive-absorption in jejunum, elimination (total body clearance), and volume of distribution. Note that simulations performed in Oral Bioavailability module ignore first-pass metabolism in liver and gut. To include the first-pass effect in simulations consider using PK Explorer module. | |||
Quantitative ''%F'' values are calculated as a ratio of AUCs after oral and intravenous administration. Also, the employed simulation model allows evaluating the dose dependence of bioavailability. | |||
<br> | <br> | ||
===Validation=== | |||
==== | ====Validation Set & Assessment Procedure==== | ||
Since the predictions are based on a mechanistic simulation model rather that formal statistical fitting to a set of data points, the validation procedure was not based on a conventional training/test set approach. Instead, a set of clinical fraction absorbed (''f<sub>a</sub>'') data together with dosage information for 28 drugs reported by Parrott & Lavé [http://www.ncbi.nlm.nih.gov/pubmed/12356420] (originating mainly from a compilation by Zhao et al. [http://www.ncbi.nlm.nih.gov/pubmed/11357178]) was used for validation purposes. | |||
The model performance was assessed in several ways: | |||
* Qualitatively, by evaluating the accuracy of three-class classification, where the compounds were categorized by their calculated extent of absorption in the following manner: | |||
** Low: ''f<sub>a</sub>'' ≤ 33% | |||
** Moderate: 33% < ''f<sub>a</sub>'' < 66% | |||
** High: ''f<sub>a</sub>'' ≥ 66% | |||
* Quantitatively, using the Residual Mean Square Error (RMSE) statistic and visual inspection of the correlation between observed and predicted ''f<sub>a</sub>'' values of the considered drugs. | |||
<br> | <br> | ||
< | |||
{| cellpadding="2" cellspacing="0" style="border-top: | ====Validation results==== | ||
|+ <b>Table | |||
The model performance for validation set compounds is demonstrated in the Table below. Notably, the software did not produce any two-class misclassification errors (when a molecule having low ''f<sub>a</sub>'' is predicted to be well-absorbed or vice versa) except pirenzepine (exp. ''f<sub>a</sub>'' = 27% at 50 mg) which is known as P-glycoprotein substrate [http://www.ncbi.nlm.nih.gov/pubmed/12438524] and its bioavailability is significantly limited by P-gp efflux. | |||
{| cellpadding="2" cellspacing="0" align="center" style="border-top:1px solid black" | |||
|+ <b>Table.</b> Qualitative and quantitative fraction absorbed predictions for the validation set of 28 drugs. | |||
|- | |||
! style="border-bottom:1px solid black; background:#EAEAEA" width="200" | Compound name | |||
! style="border-bottom:1px solid black; background:#EAEAEA" width="175" | Dose, mg | |||
! style="border-bottom:1px solid black; background:#EAEAEA" width="175" | Experimental ''f<sub>a</sub>'' (%) | |||
! style="border-bottom:1px solid black; background:#EAEAEA" width="175" | Predicted ''f<sub>a</sub>'' (%) | |||
! style="border-bottom:1px solid black; background:#EAEAEA" width="175" | Experimental class | |||
! style="border-bottom:1px solid black; background:#EAEAEA" width="175" | Predicted class | |||
|- | |||
| Acyclovir | |||
| align="center" | 350 | |||
| align="center" | 23 | |||
| align="center" | 17.9 | |||
| align="center" style="background:#FFCCCC" | Low | |||
| align="center" style="background:#FFCCCC" | Low | |||
|- | |||
| Amiloride | |||
| align="center" | 10 | |||
| align="center" | 50 | |||
| align="center" | 9.1 | |||
| align="center" style="background:#FFFFCC" | Moderate | |||
| align="center" style="background:#FFCCCC" | Low | |||
|- | |||
| Antipyrine | |||
| align="center" | 600 | |||
| align="center" | 97 | |||
| align="center" | 99.5 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |||
| Atenolol | |||
| align="center" | 50 | |||
| align="center" | 50 | |||
| align="center" | 13.9 | |||
| align="center" style="background:#FFFFCC" | Moderate | |||
| align="center" style="background:#FFCCCC" | Low | |||
|- | |||
| Carbamazepine | |||
| align="center" | 200 | |||
| align="center" | 70 | |||
| align="center" | 78 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |||
| Chloramphenicol | |||
| align="center" | 250 | |||
| align="center" | 90 | |||
| align="center" | 98.9 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |||
| Desipramine | |||
| align="center" | 150 | |||
| align="center" | 100 | |||
| align="center" | 99.2 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |||
| Diazepam | |||
| align="center" | 5 | |||
| align="center" | 100 | |||
| align="center" | 99 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |||
| Diltiazem | |||
| align="center" | 90 | |||
| align="center" | 90 | |||
| align="center" | 99.7 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |||
| Etoposide | |||
| align="center" | 350 | |||
| style="color: red;" align="center" | ''50'' | |||
| align="center" | 95.8 | |||
| align="center" style="background:#FFFFCC" | Moderate | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |||
| Furosemide | |||
| align="center" | 80 | |||
| align="center" | 61 | |||
| align="center" | 30.6 | |||
| align="center" style="background:#FFFFCC" | Moderate | |||
| align="center" style="background:#FFCCCC" | Low | |||
|- | |||
| Ganciclovir | |||
| align="center" | 75 | |||
| align="center" | 3 | |||
| align="center" | 8.9 | |||
| align="center" style="background:#FFCCCC" | Low | |||
| align="center" style="background:#FFCCCC" | Low | |||
|- | |||
| Hydrochlorothiazide | |||
| align="center" | 50 | |||
| align="center" | 69 | |||
| align="center" | 63.1 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#FFFFCC" | Moderate | |||
|- | |||
| Ketoprofen | |||
| align="center" | 75 | |||
| align="center" | 92 | |||
| align="center" | 99.4 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |||
| Metoprolol | |||
| align="center" | 100 | |||
| align="center" | 95 | |||
| align="center" | 96.9 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |||
| Naproxen | |||
| align="center" | 500 | |||
| align="center" | 99 | |||
| align="center" | 99.5 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |||
| Penicillin V | |||
| align="center" | 200 | |||
| align="center" | 38 | |||
| align="center" | 65 | |||
| align="center" style="background:#FFFFCC" | Moderate | |||
| align="center" style="background:#FFFFCC" | Moderate | |||
|- | |||
| Pirenzepine | |||
| align="center" | 50 | |||
| style="color: red;" align="center" | ''27'' | |||
| align="center" | 94 | |||
| align="center" style="background:#FFCCCC" | Low | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |- | ||
| Piroxicam | |||
| align="center" | 20 | |||
| align="center" | 100 | |||
| align="center" | 94.5 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |- | ||
| Progesterone | |||
| align="center" | 2.5 | |||
| align="center" | 100 | |||
| align="center" | 93.6 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |- | ||
| | | Propranolol | ||
| | | align="center" | 240 | ||
| align="center" | 99 | |||
| align="center" | 99.3 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |- | ||
| | | Ranitidine | ||
| align="center" | 60 | |||
| align="center" | 63 | |||
| align="center" | 51.1 | |||
| align="center" style="background:#FFFFCC" | Moderate | |||
| align="center" style="background:#FFFFCC" | Moderate | |||
|- | |- | ||
| | | Saquinavir | ||
| | | align="center" | 600 | ||
| style="color: red;" align="center" | ''30'' | |||
| | | align="center" | 48.4 | ||
| | | align="center" style="background:#FFCCCC" | Low | ||
| align="center" style="background:#FFFFCC" | Moderate | |||
|- | |- | ||
| | | Sulpiride | ||
| | | align="center" | 200 | ||
| align="center" | 44 | |||
| | | align="center" | 41.9 | ||
| align="center" style="background:#FFFFCC" | Moderate | |||
| align="center" style="background:#FFFFCC" | Moderate | |||
|- | |- | ||
| | | Terbutaline | ||
| | | align="center" | 10 | ||
| align="center" | 62 | |||
| | | align="center" | 19.5 | ||
| | | align="center" style="background:#FFFFCC" | Moderate | ||
| align="center" style="background:#FFCCCC" | Low | |||
| | |||
|- | |- | ||
| Theophylline | |||
| align="center" | 200 | |||
| align="center" | 100 | |||
| align="center" | 99.5 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |- | ||
| Verapamil | |||
| align="center" | 120 | |||
| align="center" | 100 | |||
| align="center" | 99.1 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |- | ||
| | | Warfarin | ||
| | | align="center" | 5 | ||
| align="center" | 98 | |||
| align="center" | 99.1 | |||
| align="center" style="background:#CCFFCC" | High | |||
| align="center" style="background:#CCFFCC" | High | |||
|- | |- | ||
| style="border-bottom: | | style="border-bottom:1px solid black; border-top:1px solid black;" | '''Statistics''' | ||
| style="border-bottom:1px solid black; border-top:1px solid black;" align="center" | '''No. of compounds: 28''' | |||
| style="border-bottom:1px solid black; border-top:1px solid black;" align="center" | '''RMSE: <span style="color: red;">''22%''</span><sup>*</sup>''' | |||
| style="border-bottom:1px solid black; border-top:1px solid black;" align="center" | '''RMSE: 17%<sup>**</sup>''' | |||
| style="border-bottom:1px solid black; border-top:1px solid black;" align="center" | '''Correct: 20/28 (<span style="color: red;">''71%''</span>)<sup>*</sup>''' | |||
| style="border-bottom:1px solid black; border-top:1px solid black;" align="center" | '''Correct: 20/25 (80%)<sup>**</sup>''' | |||
|- | |- | ||
|} | |} | ||
<span style="font-size:8pt"> | |||
<nowiki>*</nowiki> - P-gp substrates included<br> | |||
<nowiki>**</nowiki> - P-gp substrates excluded | |||
</span> | |||
Qualitative evaluation does not reveal the full picture, since compounds with ''f<sub>a</sub>'' values near the class boundaries would often be misclassified, even though the actual prediction would not be far off the experimental value. Therefore, quantitative predictions were also visualized in the scatter plot below: | |||
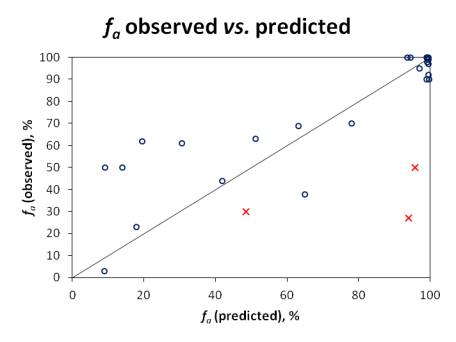
[[Image:Oral_Bioavailability_Validation_Plot.png|center|frame|'''Figure.''' Correlation between observed and predicted ''f<sub>a</sub>'' for the validation set of 28 drugs.]] | |||
| | |||
Since carrier-mediated transport effects can be reliably accounted in the simulation only if experimentally measured permeabilities are used, Pinrenzepine and two other compounds clearly affected by P-gp efflux (Saquinavir and Etoposide) were excluded from quantitative analysis - these are marked by red crosses in the scatter plot and the respective experimental values are highlighted in red in the data table. For the majority of remaining drugs, calculated values are in good agreement with clinical data, except several basic molecules (such as terbutaline, amiloride) underpredicted by more than 30%. However, one cannot disregard the possible contribution of carrier-mediated influx for these small cationic compounds. Once again, the overall RMSE of 17% was obtained with the software operating in 'pure' ''in silico'' mode, ''i.e.'', the predictions were performed using only the compounds' structure as input. On the other hand, ACD/PK Explorer module provides the potential for further improvement of prediction accuracy by offering full ''in combo'' simulation possibilities with a wide range of accepted input parameters including basic physicochemical properties (LogP and pKa), solubility, permeability, elimination rate constant, and volume of distribution. | |||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 10:04, 15 June 2017
Overview
Oral Bioavailability module predicts the fraction of the specified drug dose that reaches systemic circulation after oral administration (%F). For calculation of quantitative %F values and exploring the dose-dependence of bioavailability, Oral bioavailability module uses the same kind of absorption simulation that is implemented in ACD/PK Explorer.
Features
- Predicts %F after oral administration with the possibility to explore dose-dependence of bioavailability
- Predicts a number of endpoints that affect oral bioavailability:
- Solubility (dose/solubility ratio)
- Stability in acidic media
- Intestinal membrane permeability by passive or active transport
- Likelihood of P-gp efflux
- First pass metabolism in the liver
- Visualizes the contributions of underlying properties with traffic-lights (green = good, red = problematic) for easy interpretation
- Displays experimental %F values for up to 5 similar structures from Bioavailability DB along with literature references.
Interface
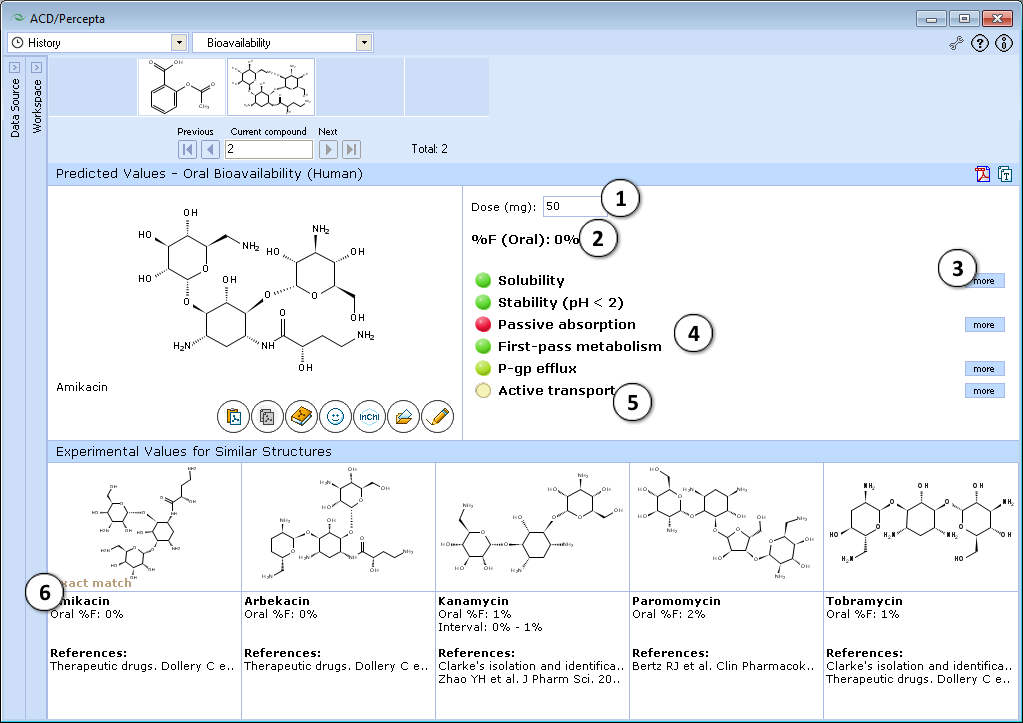
- 50 mg is the default oral drug dose used in calculations. Enter any desired value to explore the effect of dose on oral bioavailability.

- a. Click the "Undo" button to reset the specified drug dose and to recalculate %F using the default settings.
- b. Click to recalculate %F using the currently specified dose.
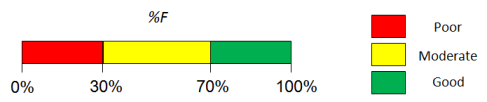
- Predicted %F (Oral) value.
- Click to see more details regarding the calculation of the particular property.
- Factors affecting oral bioavailability (see below for details).
- Hover over a title to view a screentip with a short description.
- Up to 5 similar structures in the Bioavailability DB with experimental values and references.
Traffic-lights system explanation
- Solubility in gastrointestinal tract:
- Green – good – dose/solubility ratio < 1.
- Yellow – moderate – dose/solubility ratio between 1 and 10.
- Red – poor – dose/solubility ratio > 10.
- Stability – susceptibility to acid hydrolysis in stomach:
- Red – only assigned to highly reactive compounds that decompose in stomach very quickly. Red light means that %F (oral) <10%, overriding all other predictions (i.e., do not pay attention to predicted values in this particular case).
- Passive absorption – ability to cross human intestinal membrane by passive diffusion:
- Red – intestinal passive absorption <30%. Poor bioavailability, as %F (oral) cannot exceed the extent of passive absorption.
- Green – good (>70%) passive absorption across intestinal barrier. Passive absorption does not affect %F.
- First-pass metabolism – susceptibility to metabolic transformations catalyzed by enzymes in liver and intestine:
- Red – high probability that first-pass metabolism is >50%. In this case %F (oral) is likely to be dose dependent and not to exceed 40%.
- Green – compound probably does not undergo significant first-pass metabolism.
- P-gp efflux – susceptibility to backward transport through intestinal membrane:
- Red – compound is P-glycoprotein substrate. This effect is mostly important when compound is metabolized by CYP3A4
- Active transport – susceptibility to active transport through intestinal membrane:
- Green – compound is actively transported by PepT1, ASBT or other enzymes.
- Red light never appears, as this factor can only increase %F (oral).
Technical information
Bioavailability DB
Number of compounds: 788
Main sources of experimental data:
- Reference books:
- Therapeutic Drugs, Dolery, C., Ed. 2nd Edition, Churchill Livingstone, New York, NY, 1999
- Clarke's Isolation and Identification of Drugs, Moffat, A.C., Jackson, J.V., Moss, M.S., Widdop, B., Eds. 2nd Edition, The Pharmaceutical Press, London, 1986
- Various articles from peer-reviewed scientific journals, including both detailed pharmacokinetic characterization studies (i.e., usually only several compounds per article), and larger data compilations
Simulation model
The mathematical model that is used for simulations performed by PK Explorer and Oral Bioavailability modules is based on differential equations that consider solubility in the gastrointestinal tract, passive-absorption in jejunum, elimination (total body clearance), and volume of distribution. Note that simulations performed in Oral Bioavailability module ignore first-pass metabolism in liver and gut. To include the first-pass effect in simulations consider using PK Explorer module.
Quantitative %F values are calculated as a ratio of AUCs after oral and intravenous administration. Also, the employed simulation model allows evaluating the dose dependence of bioavailability.
Validation
Validation Set & Assessment Procedure
Since the predictions are based on a mechanistic simulation model rather that formal statistical fitting to a set of data points, the validation procedure was not based on a conventional training/test set approach. Instead, a set of clinical fraction absorbed (fa) data together with dosage information for 28 drugs reported by Parrott & Lavé [1] (originating mainly from a compilation by Zhao et al. [2]) was used for validation purposes.
The model performance was assessed in several ways:
- Qualitatively, by evaluating the accuracy of three-class classification, where the compounds were categorized by their calculated extent of absorption in the following manner:
- Low: fa ≤ 33%
- Moderate: 33% < fa < 66%
- High: fa ≥ 66%
- Quantitatively, using the Residual Mean Square Error (RMSE) statistic and visual inspection of the correlation between observed and predicted fa values of the considered drugs.
Validation results
The model performance for validation set compounds is demonstrated in the Table below. Notably, the software did not produce any two-class misclassification errors (when a molecule having low fa is predicted to be well-absorbed or vice versa) except pirenzepine (exp. fa = 27% at 50 mg) which is known as P-glycoprotein substrate [3] and its bioavailability is significantly limited by P-gp efflux.
| Compound name | Dose, mg | Experimental fa (%) | Predicted fa (%) | Experimental class | Predicted class |
|---|---|---|---|---|---|
| Acyclovir | 350 | 23 | 17.9 | Low | Low |
| Amiloride | 10 | 50 | 9.1 | Moderate | Low |
| Antipyrine | 600 | 97 | 99.5 | High | High |
| Atenolol | 50 | 50 | 13.9 | Moderate | Low |
| Carbamazepine | 200 | 70 | 78 | High | High |
| Chloramphenicol | 250 | 90 | 98.9 | High | High |
| Desipramine | 150 | 100 | 99.2 | High | High |
| Diazepam | 5 | 100 | 99 | High | High |
| Diltiazem | 90 | 90 | 99.7 | High | High |
| Etoposide | 350 | 50 | 95.8 | Moderate | High |
| Furosemide | 80 | 61 | 30.6 | Moderate | Low |
| Ganciclovir | 75 | 3 | 8.9 | Low | Low |
| Hydrochlorothiazide | 50 | 69 | 63.1 | High | Moderate |
| Ketoprofen | 75 | 92 | 99.4 | High | High |
| Metoprolol | 100 | 95 | 96.9 | High | High |
| Naproxen | 500 | 99 | 99.5 | High | High |
| Penicillin V | 200 | 38 | 65 | Moderate | Moderate |
| Pirenzepine | 50 | 27 | 94 | Low | High |
| Piroxicam | 20 | 100 | 94.5 | High | High |
| Progesterone | 2.5 | 100 | 93.6 | High | High |
| Propranolol | 240 | 99 | 99.3 | High | High |
| Ranitidine | 60 | 63 | 51.1 | Moderate | Moderate |
| Saquinavir | 600 | 30 | 48.4 | Low | Moderate |
| Sulpiride | 200 | 44 | 41.9 | Moderate | Moderate |
| Terbutaline | 10 | 62 | 19.5 | Moderate | Low |
| Theophylline | 200 | 100 | 99.5 | High | High |
| Verapamil | 120 | 100 | 99.1 | High | High |
| Warfarin | 5 | 98 | 99.1 | High | High |
| Statistics | No. of compounds: 28 | RMSE: 22%* | RMSE: 17%** | Correct: 20/28 (71%)* | Correct: 20/25 (80%)** |
* - P-gp substrates included
** - P-gp substrates excluded
Qualitative evaluation does not reveal the full picture, since compounds with fa values near the class boundaries would often be misclassified, even though the actual prediction would not be far off the experimental value. Therefore, quantitative predictions were also visualized in the scatter plot below:
Since carrier-mediated transport effects can be reliably accounted in the simulation only if experimentally measured permeabilities are used, Pinrenzepine and two other compounds clearly affected by P-gp efflux (Saquinavir and Etoposide) were excluded from quantitative analysis - these are marked by red crosses in the scatter plot and the respective experimental values are highlighted in red in the data table. For the majority of remaining drugs, calculated values are in good agreement with clinical data, except several basic molecules (such as terbutaline, amiloride) underpredicted by more than 30%. However, one cannot disregard the possible contribution of carrier-mediated influx for these small cationic compounds. Once again, the overall RMSE of 17% was obtained with the software operating in 'pure' in silico mode, i.e., the predictions were performed using only the compounds' structure as input. On the other hand, ACD/PK Explorer module provides the potential for further improvement of prediction accuracy by offering full in combo simulation possibilities with a wide range of accepted input parameters including basic physicochemical properties (LogP and pKa), solubility, permeability, elimination rate constant, and volume of distribution.